Abstract
The simultaneous transmission of two lineages of the chikungunya virus (CHIKV) was discovered after the pathogen’s initial arrival in Brazil. In Oiapoque (Amapá state, north Brazil), the Asian lineage (CHIKV-Asian) was discovered, while in Bahia state, the East-Central-South-African lineage (CHIKV-ECSA) was discovered (northeast Brazil). Since then, the CHIKV-Asian lineage has been restricted to the Amazon region (mostly in the state of Amapá), whereas the ECSA lineage has expanded across the country. Despite the fact that the Asian lineage was already present in the Amazon region, the ECSA lineage brought from the northeast caused a large outbreak in the Amazonian state of Roraima (north Brazil) in 2017. Here, CHIKV spread in the Amazon region was studied by a Zika–Dengue–Chikungunya PCR assay in 824 serum samples collected between 2013 and 2016 from individuals with symptoms of viral infection in the Amapá state. We found 11 samples positive for CHIKV-Asian, and, from these samples, we were able to retrieve 10 full-length viral genomes. A comprehensive phylogenetic study revealed that nine CHIKV sequences came from a local transmission cluster related to Caribbean strains, whereas one sequence was related to sequences from the Philippines. These findings imply that CHIKV spread in different ways in Roraima and Amapá, despite the fact that both states had similar climatic circumstances and mosquito vector frequencies.
Keywords: chikungunya, metagenomics, virome, amazon region
1. Introduction
The Chikungunya virus (CHIKV) is transmitted mainly by two vectors, the Aedes aegypti and Aedes albopictus mosquitoes, which are widely distributed in different continents, including America [1,2,3], favoring its dissemination into new areas and contributing to its emergence, re-emergence, and outbreaks in different parts of the world [4]. Humans, once infected, can develop the disease, the clinical manifestations of which include symptoms such as fever, headache, nausea, fatigue, myalgia, arthralgia, and rash [5]. The recovery time ranges from weeks to years. In general, clinical severity is associated with increasing age [6]. However, there are cases in which some patients (3 to 28%) do not manifest clinical symptoms [7].
CHIKV belongs to the Togaviridae family and is classified as an Alphavirus. The genome is made up of two open reading frames (ORFs) that encode two polyproteins that are then processed into four non-structural proteins (nsP1–nsP4) and five structural proteins (capsid, E3, E2, 6K, and E1) [8]. A phylogenetic study revealed that CHIKV is divided into four genotypes or lineages: Asian, Indian Ocean (IOL), East, Central, and South Africa (ECSA), and West Africa (WA) [9].
Originally, CHIKV was isolated in 1952 on the Makonde plateau in Tanzania [10,11]. Later, it was detected in different locations in Africa, Asia, Europe, and in the Indian and Pacific oceans [12]. In South America, CHIKV was first introduced in late 2013 from the Caribbean islands [13]. Since then, it has spread to different countries in South America. In Brazil, the first detection of autochthonous CHIKV cases was confirmed in September 2014 through two independent introductions [14]. The Asian/Caribbean lineage (CHIKV-Asian), first identified in September 2014 in the Oiapoque municipality, state of Amapá, followed by the East-Central-South African (CHIKV-ECSA) lineage, which was introduced in Brazil in the city of Feira-de-Santana, state of Bahia [14,15]. Since then, several autochthonous cases have been described [5,16]. Likewise, several imported cases were reported [17,18]. In Roraima state, in the Amazon region, CHIKV-Asian was initially introduced in 2015, and a large CHIKV outbreak occurred in 2017, caused by an ECSA-lineage [18].
The Brazil–Guyana transboundary zone (ZTBG) is in the north of the South American continent, with a hot and humid climate. The ZTBG is home to four municipalities: Saint-Georges de L’Oyapoque, Camopi, and Ouanary on the Guyanese side, and Oiapoque on the Brazilian side. The municipality of Oiapoque is located 590 kilometers from Macapá, the state capital of Amapá (Brazil). The municipality of Oiapoque, with a population of 28,000 people, is ZTBG’s largest and most important commercial center, with considerable movement of people between Brazil and Guyana.
Since the arrival of the CHIKV in South America from the Caribbean Islands, French Guyana was one of the countries most affected by the epidemic [19]; promptly, the municipality of Oiapoque also suffered from the Chikungunya epidemic (with 1541 cases between 2014 and 2016), thus demonstrating that this border region is an important gateway for the emergence of new viral epidemics. In this study, we report ten near-full length CHIKV sequences detected in individuals from Amapá, north Brazil. All of these sequences are from the CHIKV-Asian lineage, and nine of them form a single monophyletic clade, implying a local transmission cluster. One of the Amapá sequences was found to be closely related to strains from the Philippines, implying that CHIKV-Asian transmission in the Amazonian region has multiple strains originated from distinct geographical locations.
2. Materials and Methods
2.1. Sample Collection
A total number of 824 serum samples from individuals presenting with symptoms compatible with arbovirus infection were obtained from the LACEN (Laboratório Central) of the state of Amapá, of which 96, 240, 283, and 205 samples were available from the years 2013, 2014, 2015, and 2016, respectively. Additionally, epidemiological data, such as the date of symptom onset (i.e., fever with headache, myalgia, arthralgia, weakness, etc.), the date of sample collection, sex, age, and municipality of residence were collected for molecular diagnostics.
2.2. Sample Processing and Quantitative Real-Time RT-PCR
A Roche MagNA Pure 2.0 automatic nucleic acid extraction machine was used to extract viral RNA from the samples (MagNA Pure LC instrument, Roche Applied Science, Indianapolis, IN, USA.). The extraction reagent kits were from Roche’s MagNA Pure LC Total Nucleic Acid Isolation Kit High Performance, Version 8, and the methodology used was that stated in the kit instructions. Each sample was extracted with a volume of 200 μL of blood plasma; if the sample did not have a total volume of 200 μL, PBS was added to the sample and the contents of the sample tube were gently stirred with a pipette. Each sample had a final elution volume of 60 μL. The samples were kept in a −80 °C freezer after extraction. After that, the samples were put through a sequence of qPCR tests. To begin, all samples were subjected to the BIORAD (Bio-Rad Laboratories, Inc.; Hercules, California) ZDC (Zika, Dengue, Chikungunya-ZDC-PCR) Multiplex qPCR Assay. The assay was carried out according to the manufacturer’s protocol, which is included with the kit, using 5 μL of extracted RNA. The samples that tested positive for the ZDC assay were sent to NGS for analysis.
2.3. Library Preparation and Next-Generation Sequencing
The preparation of the next-generation sequencing library was performed as described by da Costa [20]. To eliminate host and bacterial cellular debris, 0.3 mL of the sample was centrifuged at 12,000× g for 5 min at 8 °C and the supernatant was filtered through a 0.45 M Millipore filter (Billerica, MA, USA). The filtrate was then treated for 1.5 h at 37 °C with a mixture of nucleases to decrease nucleic acids, keeping only the infectious viral nucleic acids, which are protected from digestion by their capsids. The ZR & ZR-96 DNA/RNA kits were then used to extract total nucleic acid (Zymo Research, Irvine, CA, USA). The elution volume was 0.05 mL of nuclease-free water, according to the manufacturer’s guidelines. The SuperScript III kit (Life Technologies, Grand Island, NY, USA) was used to make the first strand of cDNA, and the Klenow FRAGMENT kit was used to make the second strand (New England Biolabs, Ipswich, MA, USA). After that, the final product was sent to Nextera XT (Illumina, San Diego, CA, USA) for library preparation. Illumina software was used to demultiplex the paired-end, 300 pb sequences generated by MiSeq. The data was then run via the Blood Systems Research Institute’s “virus discovery” pipeline on supercomputers (Deng et al., 2015). Bowtie2 was used to filter the sequences, excluding human, bacterial, and fungal sequences. SOAPdenovo2, Abyss, meta-Velvet, CAP3, Mira, and SPADES algorithms were later used to reconstruct viral genomes. BLASTx and BLASTn were used to analyze the contigs. Using Geneious R8, the quality and coverage of the entire or partial genomes were assessed (Biomatters, San Francisco, CA, USA).
2.4. Phylogenetic and Bayesian Analysis
Firstly, we submitted the sequences generated in this study to a genotyping analysis using the phylogenetic arbovirus subtyping tool, available at http://genomedetective.com/app/typingtool/chikungunya (accessed on 8 May 2022) [21]. To investigate the phylodynamics of CHIKV in the state of Amapá, we downloaded all sequences assigned as CHIKV from GenBank (n = 6232), submitted them to Genome detective, and selected only Asian and Caribbean genotype sequences out of the remaining 1034 sequences. This dataset, plus Brazilian sequences, was aligned using MAFFT [22] and edited using AliView [23]. Partial, poorly aligned, and identical sequences were removed from the dataset. A final dataset, with 257 sequences, was used for phylogenetic analysis. We estimated maximum-likelihood phylogenies in PhyML [24] by using the best-fit model of nucleotide substitution, as indicated by the jModelTest application [25]. To investigate the temporal signal in our CHIKV dataset, we regressed root-to-tip genetic distances from this ML tree against sample collection dates using TempEst v 1.5.1. Time-scaled phylogenetic trees were inferred by using the BEAST package v.1.10.4 [26]. We employed a model selection analysis using both path-sampling and stepping stone models to estimate the most appropriate model combination for Bayesian phylogenetic analysis, and the best fitting model was the TN-93 plus Gamma correction substitution model with a Bayesian skyline coalescent model. Phylogeographic analyses were applied as an asymmetric model of location transitioning coupled with the Bayesian stochastic search variable selection (BSSVS) procedure. We complemented this analysis with Markov jump estimation that counts location transitions per unit time along the tree. The Monte Carlo Markov chains ran long enough to ensure stationarity and adequate effective sample size (ESS) of >200. A final maximum clade credibility tree was generated by summarizing the results of Bayesian phylogenetic inference and viewed in FigTree software [20,27].
2.5. Epidemiological Data Compilation
Data of weekly notified CHIKV cases in Brazil and the state of Amapá were collected from Sistema de Informação de agravos de notificação-SINAN (http://portalsinan.saude.gov.br/sinan-dengue-chikungunya (accessed on 24 April 2022)) and bulletins from the Superintendence of Health Surveillance of State of Amapá (https://svs.portal.ap.gov.br/publicacoes (accessed on 24 April 2022)).
3. Results
3.1. PCR Assay
We processed 824 samples using the ZDC-PCR assay, of which 788 were negative and 36 samples (4.3%) were positive (0 from 2013, 8 from 2014, 23 from 2015, and 5 from 2016). Of the 36 positive samples, 24 were DENV (5 from 2014; 16 from 2015; and 3 from 2016), 11 were CHIKV (3 from 2014; 7 from 2015; and 1 from 2016), and 1 was ZIKV (from 2016). Partial results of this serum survey were previously published [20,27]. Regarding the 11 CHIKV positive samples, the average RT-PCR cycle threshold was 28.02 (ranging from 20.2 to 37.35), and was from patients with an average age of 32 years of age, of which the majority (67.6%) were female (Table 1).
Table 1.
Epidemiological data for the CHIKV samples.
| Sample ID | Collection Date (Y-M-D) | Age 1 | Sex | Municipalities | Cycle Threshold |
|---|---|---|---|---|---|
| 135 | 2014-10-10 | 54 | F | Macapá | 28.21 |
| 139 | 2014-10-24 | 20 | F | Macapá | 22.56 |
| 181 | 2014-10-28 | 29 | F | Macapá | 26.93 |
| 351 | 2015-01-09 | 21 | M | Macapá | 32.77 |
| 539 | 2015-02-03 | 16 | F | Laranjal do Jari | 35.61 |
| 548 | 2015-02-20 | 42 | M | Macapá | 20.64 |
| 564 | 2015-02-03 | 61 | F | Macapá | 37.35 |
| 574 * | 2015-01-29 | 30 | M | Porto Grande | NA |
| 575 | 2015-02-24 | 30 | F | Macapá | 27.93 |
| 613 | 2015-02-25 | 26 | F | Macapá | 20.2 |
| 695 | 2016-03-11 | 28 | M | Macapá | NA |
NA = Not Available; F = Female; M = Male. * From this sample we were not able to sequence the CHIKV genome; 1 Age at collection date.
3.2. Location of Sample Collection
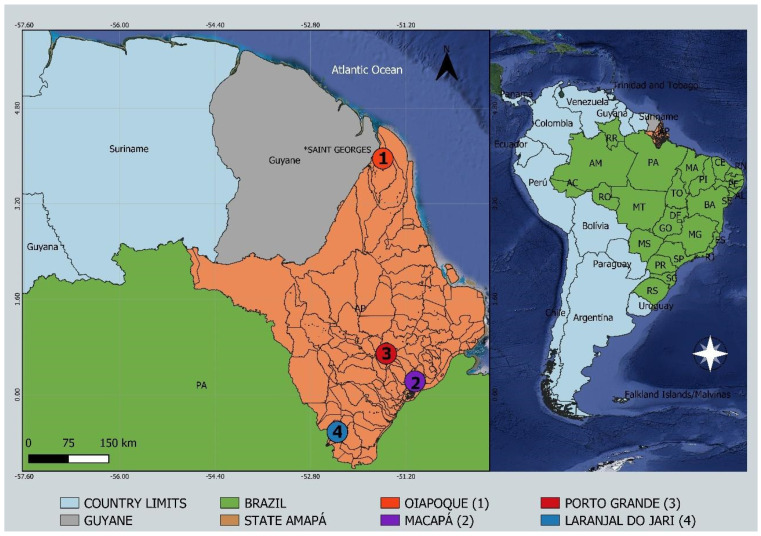
The patients lived in the cities of Macapá (capital of the state of Amapá), Laranjal do Jari, and Porto Grande Municipalities (Figure 1). At the time of sample collection, patients had the following symptoms: fever in the last seven days, pain in the muscle, localized exanthema, and cough.
Figure 1.
Study sites in the state of Amapá, northern Brazil, showing locations of sequenced cases of CHIKV-Asian outbreak. Circles indicate municipalities: (1) Oiapoque; (2) Macapá; (3) Porto Grande; (4) Laranjal do Jari.
3.3. Next Generation Sequencing and Genotyping Tree
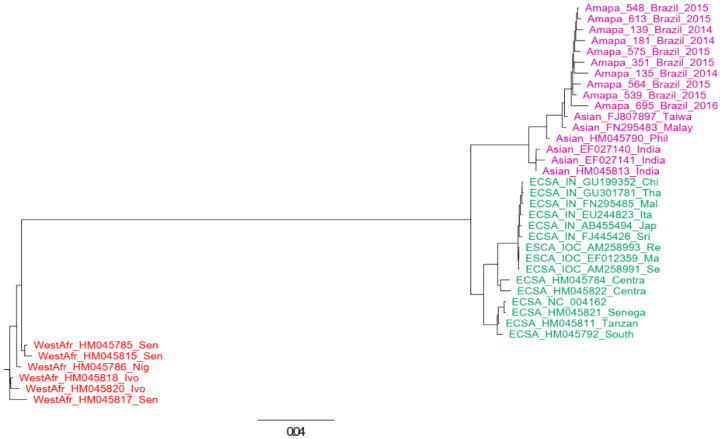
From the CHIKV samples, we were able to generate 10 complete or near-complete genome sequences and deposited them in GenBank at NCBI access numbers OL343608–OL343617. The identification of CHIKV genotypes was performed using phylogenetic analysis of full-length genome datasets and using an online tool (http://genomedetective.com/app/typingtool/chikungunya (accessed on 8 May 2022)). We also inferred a maximum likelihood tree that includes more CHIKV-Asian references and variants from other countries to give more support to the classification of our sequences (tree not shown). Both approaches indicated that all sequences generated in this study belong to the Asian lineage (Figure 2).
Figure 2.
Genotyping tree of CHIKV. The tree was constructed using complete genomes of CHIKV. The maximum likelihood approach was used and the model GTR was assumed in this inference. Sequences of Asian lineage are indicated in magenta, those of West African lineage are in red, and those of ECSA are indicated in green. The horizontal bar indicates the nucleotide substitution per base.
3.4. Phylogenetic Analysis
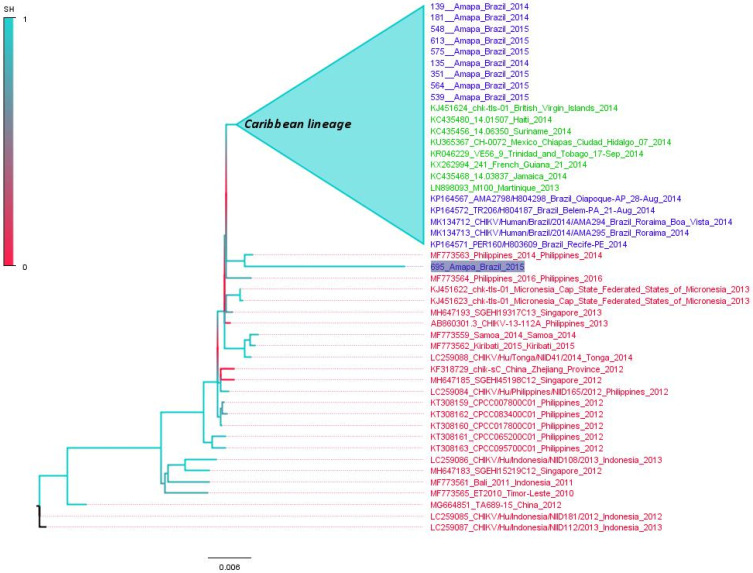
We applied the maximum likelihood (ML) criterion to construct phylogenetic trees of complete genomes of the CHIKV-Asian lineage using sequences from South Asia, the Caribbean, and the American continent. The aim here is to understand the relatedness of Brazilian sequences to CHIKV-Asian lineages. Initially, we used 257 CHIKV-Asian lineage sequences to construct a phylogenetic tree (Figure S1, Supplementary Materials). This ML tree indicates that all sequences from the Caribbean and Americas are in a monophyletic clade previously designated the Caribbean lineage. The Caribbean lineage is related to Asian sequences, particularly strains from the Philippines detected in 2014 and 2016 (GenBank accession numbers MF773563 and MF773564, respectively). This lineage also includes one sequence from the 2015 strain from French Polynesia (KR559473) [28]. In addition, all Brazilian sequences are in the Caribbean clade, with the exception of one Brazilian sequence (695_Amapa_Brazil_2016) which is related to the sequence MF773564 from the Philippines. The clade formed by the sequences and MF773564 is at the base of the Caribbean lineage. For illustrative purposes, we also constructed a small version of the ML tree with 99 sequences (Figure 3), including all sequences from South Asian/Oceania (indicated in red in the tree), all Brazilian sequences (blue in the tree) and some Caribbean references (indicated in green in the tree). This tree equally shows the monophyletic pattern of Brazilian sequences of the Caribbean lineage that are grouped into a single clade with high statistical support (0.99). In addition, the tree indicates that the sequence 695_Amapa_Brazil_2016 is not in the Caribbean clade, and is related to one sequence from the Philippines in 2014.
Figure 3.
Maximum likelihood tree of CHIKV-Asian lineage. The unrooted tree was constructed using reference genomes of CHIKV-Asian lineage. Sequences from Brazil are indicated in blue. Sequences from Caribbean countries are in green color and sequences from South Asia/Oceania are in red color. The cluster composed by Caribbean sequences plus almost all Brazilian sequences is labeled. The Brazilian sequence related with a Philippine sequence is highlighted. The branch support is indicated by a color scale of 0 to 1, and is based on the Shimodaira–Hasegawa-like test. The tree was inferred using the TN-93 model plus gamma correction. Horizontal bar indicates the nucleotide substitution per base. For better visualization of the tree some sequences were collapsed (blue triangles).
3.5. Polyphyletic versus Monophyletic Pattern of CHIKV from Amapá
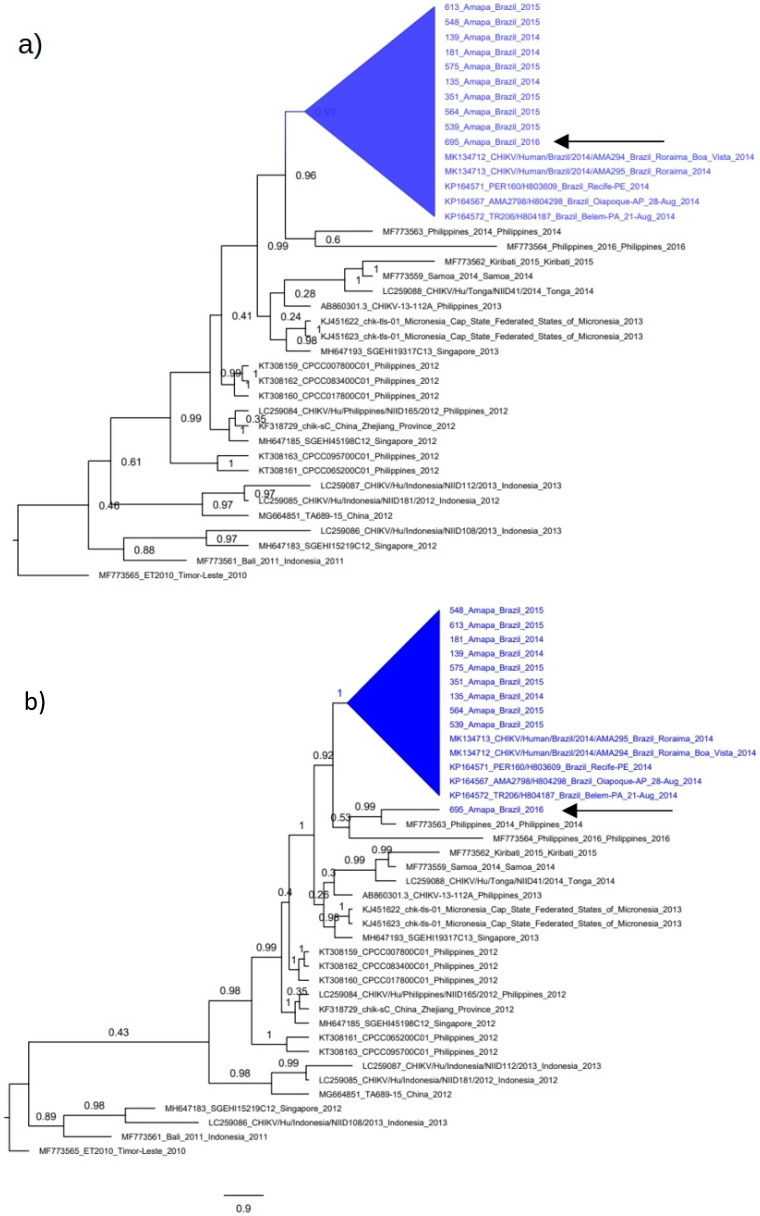
We applied a maximum likelihood hypothesis test (Shimodaira–Kishino test) to evaluate the monophyletic versus the polyphyletic pattern of our CHIKV sequences from Amapá because sequences of the Caribbean lineage have reduced diversity (less than 1% of nucleotide divergence). This low divergence, besides producing near zero branch lengths, can also impact on the grouping pattern of ML trees. To provide more support to the circulation of distinct CHIKV variants, we constructed one tree using a coalescent approach assuming that all sequences from Amapá were monophyletic, including the sequence 695_Amapa, (Figure 4a), and tested against a coalescent tree in which the sequences 695_Amapa in not monophyletic (Figure 4b). The Shimodaira–Kishino test indicated that the polyphyletic tree (Figure 4b) is the most likely tree because it has the best-log likelihood compared with the alternative monophyletic tree (i.e., 21,146.93 and 21,121.1, respectively). These results indicate that the 695_Amapa is not closely related to sequences of the Caribbean lineage.
Figure 4.
Monophyletic versus polyphyletic tree of CHIKV-Asian in Amapá. The coalescent trees were constructed using the complete genome of Brazilian CHIKV plus South Asian/Oceania references. The Bayesian approach was used and trees were constructed assuming the constant coalescent model plus the TN-93 evolutionary nucleotide model. The horizontal scale bar indicates nucleotide substitutions per site. Monophyletic clades are shown in the blue triangles in both trees. (a) Coalescent tree constructed assuming that the sequence 695_Amapa (indicated by arrow) is monophyletic. (b) Coalescent tree constructed assuming that the sequence 695_Amapa (indicated by arrow) is not within the monophyletic formed by the Brazilian sequences.
3.6. Time-Scaled Tree
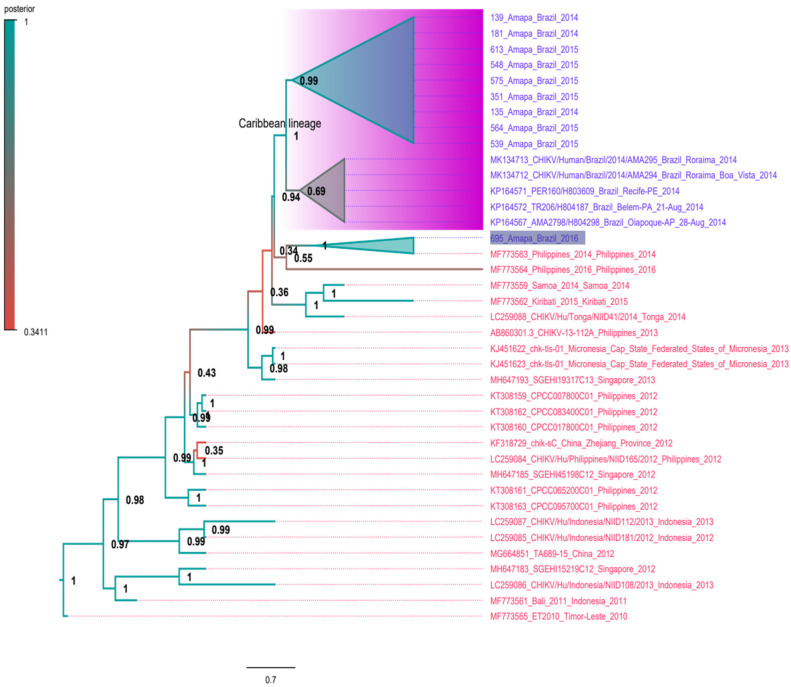
We applied a Bayesian coalescent approach to better understand the temporal pattern and the topology of trees of the CHIKV-Asian lineage. Initially, the linear regression of root-to-tip genetic distance against the sampling date from our dataset revealed a sufficient temporal signal (r2 = 0.79, Supplementary Figure S2). Next, we used a constant population size coalescent model to infer a tree in which no CHIKV of the Caribbean-lineage sequences were included besides the Brazilian sequences (Figure 5). The estimated substitution rate of the CHIKV-Asian lineage is evolving at 6.7 × 10−4 substitutions per site per year. This time-scaled Maximum clade credibility tree (MCC tree) shows that nearly all Brazilian CHIKV sequences grouped in a single clade highly the posterior probability (PP = 0.99). The exception was the sequence 695_Amapa which was grouped with sequences from the Philippines. We also used a model (Skygrid) to evaluate the fluctuation of the effective population size over time. These results indicate that CHIKV was introduced in the Caribbean region in early 2013 from South Asian variants (probably by Philippian variants) (PP= 1.0). Following this introduction, the number of infections increased steadily (Supplementary Figure S3).
Figure 5.
Time-scaled tree of CHIKV-Asian. The tree was constructed using complete genomes of CHIKV. The coalescent constant population size approach was used and the model TN-93 was assumed in this inference. Sequences of Asian/Oceania are indicated in red color and Brazilian sequences are in blue color. The Caribbean lineage is also indicated in the magenta rectangle. The sequence 695_Amapá is highlighted. Numbers in the nodes indicate the posterior probability. The horizontal bar indicates the nucleotide substitution per base.
The MCC tree also showed that the Amapá sequences were grouped into different clades, suggesting the introduction of this genotype in Amapá occurred on at least two occasions in the middle of 2014. In addition, the tree indicates that the sequence 695_Amapa is at the base of the Caribbean lineage (Figure 5).
The presence of non-synonymous substitutions already described as potential related to vector adaptability was investigated among sequences from Amapá. We observed that all 10 Amapá sequences do not carry either the substitutions A226V (E1 protein) or the L210Q (E2 protein) associated with increased CHIKV transmission in A. albopictus mosquitos. On the other hand, other substitutions related to the vector adaptability, such as T98A, A377V, M407L (E1 protein), G60D, and A103T (E2 protein), were present, as shown in Table 2.
Table 2.
Non-synonymous substitutions observed among the 10 CHIKV genomes sequenced in this study.
| Gene | Substitution | Amapá Sequences | Function |
|---|---|---|---|
| E1 | L19M | Yes | Unknow |
| T98A | Yes | Enhanced vector adaptability of A226V | |
| A226V | No | Increased infectivity, transmission, and dissemination in A. Albopictus | |
| E2 | L210Q | No | Enhanced disseminated infection in A. albopictus and fitness increment of A226V variant |
| V226 | Yes * | Unknow | |
| V367A | Yes | Unknow | |
| V384M | Yes | Unknow | |
| nsP1 | R364K | Yes | Unknow |
| Capsid | V54A | Yes | Unknow |
| 6k | L20 | Yes * | Unknow |
* Residues shared between 695_Amapa and sequences from South Asian/Oceania.
4. Discussion
The virus has spread across the region due to the widespread movement of individuals across the Brazil–Guyana border, notably for commercial reasons, as well as the region’s vulnerable health system. Unauthorized logging and mining in the Oiapoque River basin have expanded human activity in the forest region, leading to the establishment of new communities near the municipality of Oiapoque. Brazilian miners’ ventures deep into the forest have polluted the ecosystem with heavy metals used in gold mining, resulting in increased migratory movements between neighboring countries [29].
This uncontrolled flow of people creates a unique set of challenges for local health services since these highly mobile individuals are hard to track, making it difficult to assess resource needs and plan measures at the local level [30,31]. DENV-4 [32], DENV-1 [27], and CHIKV [14] are only a few examples of viral variations that have recently been found in the Brazilian Amazon.
To better understand CHIKV evolution in the state of Amapá, we sequenced ten full-length genomes and performed a phylogenetic analysis. Almost all CHIKV sequences from Macapá and Laranjal do Jari municipalities clustered in a monophyletic phylogroup, with one sequence from Macapá municipality (695 Amapá) showing a unique clustering pattern linked to a sequence reported in the Philippines in 2014 (Genbank ID: MF773563) [28].
Furthermore, the 695 Amapá and MF773563 sequence clusters are found near the base of the Caribbean lineage. With approximately 16,000 people infected between 2014 and 2015, French Guyana became the first South American country to declare CHIKV infections autochthonous in 2014. It has been claimed that South Pacific strains gave rise to the Caribbean lineage [33,34]. More recently, it has been proposed that the Caribbean lineage was imported either directly or indirectly from Southeast Asia [28]. This possibility is strengthened by our phylogenetic research. Furthermore, we discovered one Amapá sequence that, along with the Philippian strains, forms the foundation of the Caribbean lineage’s phylloclade.
It’s worth noting that, when compared to Asian/Oceanic comparisons, all Caribbean sequences include two conventional substitutions: V226A in the E2 gene and L20M in the 6K gene. Some alterations have been shown to have an effect on CHIKV fitness, mostly by interfering with the virus’s ability to transmit. In the Indian Ocean outbreak, for example, the CHIKV E2 glycoprotein substitution A226V has been linked to increased transmission by Aedes albopictus, which is thought to have the lowest vector capacity for CHIKV transmissibility than Aedes aegypti [35].
This replacement has also been implicated in European outbreaks involving Aedes albopictus [36]. L210Q arose in IOL strains in India and was linked to greater CHIKV transmission by the Aedes albopictus vector [37], while V226A boosted viral dissemination in Aedes aegypti but not in Aedes albopictus [38,39].
To our knowledge, none of the Brazilian strains (Asian or ECSA lineages) contain the substitutions A226V in the E1 glycoprotein or L210Q in the E2 glycoprotein. On the other hand, the Amapá sequences, as well as other Asian strains, have threonine (T) in position 98 of the E1 glycoprotein, which limits their infectivity in Ae. albopictus through the substitution A226V [40]. In particular, the sequence 695_Amapa has the residues V226 in the E2 gene and L20 in the 6K gene, likewise the sequences from South Asian/Oceania. Moreover, we found six amino acid changes in Amapá sequences (nsP1, capsid, E1, and E2 proteins), none of which have been described previously. The capsid, E2, and E1 glycoproteins are responsible for recognizing the host-receptor and entry into the cell [41,42], and they are widely used for vaccine development and serodiagnostic assays [43]. These substitutions in the E1 and E2 proteins could cause viral evasion and potentially allow host-switching of the CHIKV-Asian lineage in the Amazon region [44].
On the American continent, the CHIKV-Asian lineage is widely distributed, while the CHIKV-ECSA lineage is restricted only to Brazil, Paraguay, and Haiti [45,46]. In contrast, in Brazil, the CHIKV-Asian lineage was limited to a small number of cases and was geographically restricted (predominantly in the state of Amapá), whereas the ECSA lineage was widespread [20,47]. It has been shown that CHIKV-Asian is less capable of accumulating mutations that facilitate their transmission through vectors when compared to the ECSA lineage [48]. In Roraima, a state in the north of Brazil that borders Venezuela, the CHIKV-Asian lineage was introduced in 2015 from northeastern Brazil, and a large outbreak in 2017 was caused by the ECSA genotype that gradually replaced the original CHIKV strains [18]. Although Roraima and Amapá have very similar climate conditions, the CHIKV-Asian lineage continued to be predominant in the state of Amapá. It is important to mention that Ae. aegypti and Ae. albopictus are endemic in the Amazon region [49]. In addition, the dissemination of CHIKV-Asian by Ae. albopictus might be restricted due to the genetic background of the virus [39]. This pattern of CHIKV strain replacements in the Amazon region is probably affected by multiple factors such as the ecological community, human behavior, and the genetics of the virus.
The modest number of CHIKV sequences we were able to recover from the samples was perhaps the most significant limitation of our research. This has restricted our ability to examine the genetic diversity of CHIKV in depth. Multidisciplinary research will be required to understand the key factors that contribute to the transmission dynamics of CHIKV in the Amazon.
5. Conclusions
Despite the low frequency of CHIKV in our samples, we observed various strains circulating in the Amapá, indicating that CHIKV-Asian transmission in the Amazonian region originated from different geographical locations.
Acknowledgments
We thank Luciano Monteiro da Silva for the administrative support. Our thanks also to the Sage Science Inc., MP Biomedicals Inc., and Zymo Research Corporation, for the donation of reagents. We also thank Pró-reitoria de pesquisa e pós-graduação of UFPA for supporting the publication costs.
Supplementary Materials
The following supporting information can be downloaded at: https://www.mdpi.com/article/10.3390/v14071445/s1, Figure S1: Maximum likelihood tree of CHIKV-Asian lineage; Figure S2: Linear regression analysis; Figure S3: Skygrid demographic of CHIKV.
Author Contributions
Specimen collection and sample screening, D.E.G., F.J.C.M., V.S.M., M.O.d.S.R., E.L.L.A. and A.C.d.C.; conceptualization, D.E.G., E.D., É.L. and A.C.d.C.; sequencing and contig analysis, G.d.O.R., D.E.G., X.D., E.D., É.L. and A.C.d.C.; data investigation, G.d.O.R., F.V., D.E.G., E.d.S.F.R., E.S.D.R., F.J.C.M., V.S.M., M.O.d.S.R., E.L.L.A., X.D., E.D., É.L. and A.C.d.C.; writing original draft, G.d.O.R., F.V., E.S.D.R., É.L. and A.C.d.C.; manuscript editing, R.P.P. and V.S.R.; supervision, E.D., É.L. and A.C.d.C. All authors have read and agreed to the published version of the manuscript. All authors critically revised the manuscript for intellectual content and approved the final version.
Institutional Review Board Statement
All procedures performed in studies involving human participants were in accordance with the ethical standards of the institutional and/or national research committee and with the 1964 Helsinki Declaration and its later amendments or comparable ethical standards. Informed consent was obtained from adult individuals and from all parents or guardians of children participants involved in the study. Ethics Committee approval was granted by the Faculdade de Medicina da Universidade de São Paulo (CAAE: 53153916.7.0000.0065).
Informed Consent Statement
Written informed consent has been obtained from the patient(s) to publish this paper.
Conflicts of Interest
The authors declare no conflict of interest.
Funding Statement
É.L. is supported by Conselho Nacional de Desenvolvimento Científico e Tecnológico (CNPq #302677/2019-4). G.d.O.R. is supported by a scholarship provided by the Coordenação de Aperfeiçoamento de Pessoal de Nível Superior—Brasil (CAPES). A.C.d.C. is supported by a scholarship from HCFMUSP with funds donated by NUBANK under the #HCCOMVIDA scheme. V.S.M. is supported by a scholarship provided by the Fundação de Amparo à Pesquisa do Estado de São Paulo (FAPESP #2019/21706-5).
Footnotes
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations.
References
- 1.Costa-da-Silva A.L., Ioshino R.S., Petersen V., Lima A.F., dos Passos Cunha M., Wiley M.R., Ladner J.T., Prieto K., Palacios G., Costa D.D., et al. First Report of Naturally Infected Aedes aegypti with Chikungunya Virus Genotype ECSA in the Americas. PLoS Negl. Trop. Dis. 2017;11:e0005630. doi: 10.1371/journal.pntd.0005630. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Eskildsen G.A., Rovira J.R., Smith O., Miller M.J., Bennett K.L., McMillan W.O., Loaiza J. Maternal Invasion History of Aedes aegypti and Aedes albopictus into the Isthmus of Panama: Implications for the Control of Emergent Viral Disease Agents. PLoS ONE. 2018;13:e0194874. doi: 10.1371/journal.pone.0194874. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Vega-Rúa A., Zouache K., Girod R., Failloux A.-B., Lourenço-de-Oliveira R. High Level of Vector Competence of Aedes aegypti and Aedes albopictus from Ten American Countries as a Crucial Factor in the Spread of Chikungunya Virus. J. Virol. 2014;88:6294–6306. doi: 10.1128/JVI.00370-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Xavier J., Fonseca V., Bezerra J.F., do Monte Alves M., Mares-Guia M.A., Claro I.M., de Jesus R., Adelino T., Araújo E., Cavalcante K.R.L.J., et al. Chikungunya Virus ECSA Lineage Reintroduction in the Northeasternmost Region of Brazil. Int. J. Infect. Dis. 2021;105:120–123. doi: 10.1016/j.ijid.2021.01.026. [DOI] [PubMed] [Google Scholar]
- 5.Monteiro J.D., Valverde J.G., Morais I.C., de Medeiros Souza C.R., Fagundes Neto J.C., de Melo M.F., Nascimento Y.M., Alves B.E.B., de Medeiros L.G., Pereira H.W.B., et al. Epidemiologic and Clinical Investigations during a Chikungunya Outbreak in Rio Grande Do Norte State, Brazil. PLoS ONE. 2020;15:e0241799. doi: 10.1371/journal.pone.0241799. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Elsinga J., Gerstenbluth I., van der Ploeg S., Halabi Y., Lourents N.T., Burgerhof J.G., van der Veen H.T., Bailey A., Grobusch M.P., Tami A. Long-Term Chikungunya Sequelae in Curaçao: Burden, Determinants, and a Novel Classification Tool. J. Infect. Dis. 2017;216:573–581. doi: 10.1093/infdis/jix312. [DOI] [PubMed] [Google Scholar]
- 7.Yactayo S., Staples J.E., Millot V., Cibrelus L., Ramon-Pardo P. Epidemiology of Chikungunya in the Americas. J. Infect. Dis. 2016;214:S441–S445. doi: 10.1093/infdis/jiw390. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Simizu B., Yamamoto K., Hashimoto K., Ogata T. Structural Proteins of Chikungunya Virus. J. Virol. 1984;51:254–258. doi: 10.1128/jvi.51.1.254-258.1984. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Powers A.M., Brault A.C., Tesh R.B., Weaver S.C. Re-Emergence of Chikungunya and O’nyong-Nyong Viruses: Evidence for Distinct Geographical Lineages and Distant Evolutionary Relationships. Microbiology. 2000;81:471–479. doi: 10.1099/0022-1317-81-2-471. [DOI] [PubMed] [Google Scholar]
- 10.Robinson M.C. An Epidemic of Virus Disease in Southern Province, Tanganyika Territory, in 1952–1953. I. Clinical Features. Trans. R. Soc. Trop. Med. Hyg. 1955;49:28–32. doi: 10.1016/0035-9203(55)90080-8. [DOI] [PubMed] [Google Scholar]
- 11.Ross R.W. The Newala Epidemic: III. The Virus: Isolation, Pathogenic Properties and Relationship to the Epidemic. J. Hyg. 1956;54:177–191. doi: 10.1017/S0022172400044442. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Onyango C., Breiman R.F., Ofula V., Bedno S., Konongoi L.S., Burke H., Konde J., Sergon K., Sang R., Dumilla A.M., et al. Seroprevalence of Chikungunya Virus (CHIKV) Infection on Lamu Island, Kenya, October 2004. Am. J. Trop. Med. Hyg. 2008;78:333–337. doi: 10.4269/ajtmh.2008.78.333. [DOI] [PubMed] [Google Scholar]
- 13.Gallian P., Leparc-Goffart I., Richard P., Maire F., Flusin O., Djoudi R., Chiaroni J., Charrel R., Tiberghien P., de Lamballerie X. Epidemiology of Chikungunya Virus Outbreaks in Guadeloupe and Martinique, 2014: An Observational Study in Volunteer Blood Donors. PLoS Negl. Trop. Dis. 2017;11:e0005254. doi: 10.1371/journal.pntd.0005254. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Nunes M.R.T., Faria N.R., de Vasconcelos J.M., Golding N., Kraemer M.U., de Oliveira L.F., do Socorro da Silva Azevedo R., da Silva D.E.A., da Silva E.V.P., da Silva S.P., et al. Emergence and Potential for Spread of Chikungunya Virus in Brazil. BMC Med. 2015;13:102. doi: 10.1186/s12916-015-0348-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Machado L.C., de Morais-Sobral M.C., de Lima Campos T., Pereira M.R., de Fátima Pessoa Militão de Albuquerque M., Gilbert C., Franca R.F.O., Wallau G.L. Genome Sequencing Reveals Coinfection by Multiple Chikungunya Virus Genotypes in a Recent Outbreak in Brazil. PLoS Negl. Trop. Dis. 2019;13:e0007332. doi: 10.1371/journal.pntd.0007332. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Rodrigues A.M., Souza R.R.M., dos Santos Fonseca L.M., de Araújo Rolo C., Carvalho R.H., Sardi S.I., Campos G.S. Genomic Surveillance of the Chikungunya Virus (CHIKV) in Northeast Brazil after the First Outbreak in 2014. Rev. Soc. Bras. Med. Trop. 2020;53:e20190583. doi: 10.1590/0037-8682-0583-2019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Conteville L.C., Zanella L., Marín M.A., de Filippis A.M.B., Nogueira R.M.R., Vicente A.C.P., de Mendonça M.C.L. Phylogenetic Analyses of Chikungunya Virus among Travelers in Rio de Janeiro, Brazil, 2014–2015. Mem. Inst. Oswaldo Cruz. 2016;111:347–348. doi: 10.1590/0074-02760160004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Naveca F.G., Claro I., Giovanetti M., de Jesus J.G., Xavier J., de Melo Iani F.C., do Nascimento V.A., de Souza V.C., Silveira P.P., Lourenço J., et al. Genomic, Epidemiological and Digital Surveillance of Chikungunya Virus in the Brazilian Amazon. PLoS Negl. Trop. Dis. 2019;13:e0007065. doi: 10.1371/journal.pntd.0007065. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Van Bortel W., Dorleans F., Rosine J., Blateau A., Rousset D., Matheus S., Leparc-Goffart I., Flusin O., Prat C.M., Césaire R., et al. Chikungunya Outbreak in the Caribbean Region, December 2013 to March 2014, and the Significance for Europe. Eurosurveillance. 2014;19:20759. doi: 10.2807/1560-7917.ES2014.19.13.20759. [DOI] [PubMed] [Google Scholar]
- 20.Charlys da Costa A., Thézé J., Komninakis S.C.V., Sanz-Duro R.L., Felinto M.R.L., Moura L.C.C., Barroso I.M.d.O., Santos L.E.C., Nunes M.A.d.L., Moura A.A., et al. Spread of Chikungunya Virus East/Central/South African Genotype in Northeast Brazil. Emerg. Infect. Dis. 2017;23:1742–1744. doi: 10.3201/eid2310.170307. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Fonseca V., Libin P.J.K., Theys K., Faria N.R., Nunes M.R.T., Restovic M.I., Freire M., Giovanetti M., Cuypers L., Nowé A., et al. A Computational Method for the Identification of Dengue, Zika and Chikungunya Virus Species and Genotypes. PLoS Negl. Trop. Dis. 2019;13:e0007231. doi: 10.1371/journal.pntd.0007231. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Katoh K., Misawa K., Kuma K., Miyata T. MAFFT: A Novel Method for Rapid Multiple Sequence Alignment Based on Fast Fourier Transform. Nucleic Acids Res. 2002;30:3059–3066. doi: 10.1093/nar/gkf436. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Larsson A. AliView: A Fast and Lightweight Alignment Viewer and Editor for Large Datasets. Bioinformatics. 2014;30:3276–3278. doi: 10.1093/bioinformatics/btu531. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Guindon S., Dufayard J.-F., Lefort V., Anisimova M., Hordijk W., Gascuel O. New Algorithms and Methods to Estimate Maximum-Likelihood Phylogenies: Assessing the Performance of PhyML 3.0. Syst. Biol. 2010;59:307–321. doi: 10.1093/sysbio/syq010. [DOI] [PubMed] [Google Scholar]
- 25.Posada D. JModelTest: Phylogenetic Model Averaging. Mol. Biol. Evol. 2008;25:1253–1256. doi: 10.1093/molbev/msn083. [DOI] [PubMed] [Google Scholar]
- 26.Drummond A.J., Rambaut A. BEAST: Bayesian Evolutionary Analysis by Sampling Trees. BMC Evol. Biol. 2007;7:214. doi: 10.1186/1471-2148-7-214. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Ribeiro G.d.O., Gill D.E., Ribeiro E.S.D., Monteiro F.J.C., Morais V.S., Marcatti R., Rego M.O.d.S., Araújo E.L.L., Witkin S.S., Villanova F., et al. Adaptive Evolution of New Variants of Dengue Virus Serotype 1 Genotype V Circulating in the Brazilian Amazon. Viruses. 2021;13:689. doi: 10.3390/v13040689. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Pyke A.T., Moore P.R., McMahon J. New Insights into Chikungunya Virus Emergence and Spread from Southeast Asia. Emerg. Microbes Infect. 2018;7:26. doi: 10.1038/s41426-018-0024-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Dezécache C., Faure E., Gond V., Salles J.-M., Vieilledent G., Hérault B. Gold-Rush in a Forested El Dorado: Deforestation Leakages and the Need for Regional Cooperation. Environ. Res. Lett. 2017;12:034013. doi: 10.1088/1748-9326/aa6082. [DOI] [Google Scholar]
- 30.Peiter P.C. Condiciones de Vida, Situación de la Salud y Disponibilidad de Servicios de Salud en la Frontera de Brasil: Un Enfoque Geográfico. Cad. Saúde Pública. 2007;23:S237–S250. doi: 10.1590/S0102-311X2007001400013. [DOI] [PubMed] [Google Scholar]
- 31.Seid M., Castañeda D., Mize R., Zivkovic M., Varni J.W. Crossing the Border for Health Care: Access and Primary Care Characteristics for Young Children of Latino Farm Workers along the US-Mexico Border. Ambul. Pediatr. 2003;3:121–130. doi: 10.1367/1539-4409(2003)003<0121:CTBFHC>2.0.CO;2. [DOI] [PubMed] [Google Scholar]
- 32.Nunes M.R.T., Faria N.R., Vasconcelos H.B., Medeiros D.B.d.A., Silva de Lima C.P., Carvalho V.L., Pinto da Silva E.V., Cardoso J.F., Sousa E.C., Nunes K.N.B., et al. Phylogeography of Dengue Virus Serotype 4, Brazil, 2010–2011. Emerg. Infect. Dis. 2012;18:1858–1864. doi: 10.3201/eid1811.120217. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Sahadeo N.S.D., Allicock O.M., De Salazar P.M., Auguste A.J., Widen S., Olowokure B., Gutierrez C., Valadere A.M., Polson-Edwards K., Weaver S.C., et al. Understanding the Evolution and Spread of Chikungunya Virus in the Americas Using Complete Genome Sequences. Virus Evol. 2017;3:vex010. doi: 10.1093/ve/vex010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Chen R., Puri V., Fedorova N., Lin D., Hari K.L., Jain R., Rodas J.D., Das S.R., Shabman R.S., Weaver S.C. Comprehensive Genome Scale Phylogenetic Study Provides New Insights on the Global Expansion of Chikungunya Virus. J. Virol. 2016;90:10600–10611. doi: 10.1128/JVI.01166-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Tsetsarkin K.A., Vanlandingham D.L., McGee C.E., Higgs S. A Single Mutation in Chikungunya Virus Affects Vector Specificity and Epidemic Potential. PLoS Pathog. 2007;3:e201. doi: 10.1371/journal.ppat.0030201. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Severini F., Boccolini D., Fortuna C., Di Luca M., Toma L., Amendola A., Benedetti E., Minelli G., Romi R., Venturi G., et al. Vector Competence of Italian Aedes albopictus Populations for the Chikungunya Virus (E1-226V) PLoS Negl. Trop. Dis. 2018;12:e0006435. doi: 10.1371/journal.pntd.0006435. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Tsetsarkin K.A., Weaver S.C. Sequential Adaptive Mutations Enhance Efficient Vector Switching by Chikungunya Virus and Its Epidemic Emergence. PLoS Pathog. 2011;7:e1002412. doi: 10.1371/journal.ppat.1002412. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Agarwal A., Sharma A.K., Sukumaran D., Parida M., Dash P.K. Two Novel Epistatic Mutations (E1:K211E and E2:V264A) in Structural Proteins of Chikungunya Virus Enhance Fitness in Aedes aegypti. Virology. 2016;497:59–68. doi: 10.1016/j.virol.2016.06.025. [DOI] [PubMed] [Google Scholar]
- 39.Chen R., Plante J.A., Plante K.S., Yun R., Shinde D., Liu J., Haller S., Mukhopadhyay S., Weaver S.C. Lineage Divergence and Vector-Specific Adaptation Have Driven Chikungunya Virus onto Multiple Adaptive Landscapes. mBio. 2021;12:e02738-21. doi: 10.1128/mBio.02738-21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Zouache K., Failloux A.-B. Insect–Pathogen Interactions: Contribution of Viral Adaptation to the Emergence of Vector-Borne Diseases, the Example of Chikungunya. Curr. Opin. Insect Sci. 2015;10:14–21. doi: 10.1016/j.cois.2015.04.010. [DOI] [PubMed] [Google Scholar]
- 41.Chua C.-L., Sam I.-C., Chiam C.-W., Chan Y.-F. The Neutralizing Role of IgM during Early Chikungunya Virus Infection. PLoS ONE. 2017;12:e0171989. doi: 10.1371/journal.pone.0171989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Zhang R., Kim A.S., Fox J.M., Nair S., Basore K., Klimstra W.B., Rimkunas R., Fong R.H., Lin H., Poddar S., et al. Mxra8 Is a Receptor for Multiple Arthritogenic Alphaviruses. Nature. 2018;557:570–574. doi: 10.1038/s41586-018-0121-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Fumagalli M.J., de Souza W.M., Espósito D.L.A., Silva A., Romeiro M.F., Martinez E.Z., da Fonseca B.A.L., Figueiredo L.T.M. Enzyme-Linked Immunosorbent Assay Using Recombinant Envelope Protein 2 Antigen for Diagnosis of Chikungunya Virus. Virol. J. 2018;15:112. doi: 10.1186/s12985-018-1028-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Cook J.D., Lee J.E. The Secret Life of Viral Entry Glycoproteins: Moonlighting in Immune Evasion. PLoS Pathog. 2013;9:e1003258. doi: 10.1371/journal.ppat.1003258. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Gräf T., Vazquez C., Giovanetti M., de Bruycker-Nogueira F., Fonseca V., Claro I.M., de Jesus J.G., Gómez A., Xavier J., de Mendonça M.C.L., et al. Epidemiologic History and Genetic Diversity Origins of Chikungunya and Dengue Viruses, Paraguay. Emerg. Infect. Dis. 2021;27:1393–1404. doi: 10.3201/eid2705.204244. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.White S.K., Mavian C., Salemi M., Morris J.G., Elbadry M.A., Okech B.A., Lednicky J.A., Dunford J.C. A New “American” Subgroup of African-Lineage Chikungunya Virus Detected in and Isolated from Mosquitoes Collected in Haiti, 2016. PLoS ONE. 2018;13:e0196857. doi: 10.1371/journal.pone.0196857. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.De Souza T., Ribeiro E., Corrêa V., Damasco P., Santos C., de Bruycker-Nogueira F., Chouin-Carneiro T., Faria N., Nunes P., Heringer M., et al. Following in the Footsteps of the Chikungunya Virus in Brazil: The First Autochthonous Cases in Amapá in 2014 and Its Emergence in Rio de Janeiro during 2016. Viruses. 2018;10:623. doi: 10.3390/v10110623. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Weaver S.C. Arrival of Chikungunya Virus in the New World: Prospects for Spread and Impact on Public Health. PLoS Negl. Trop. Dis. 2014;8:e2921. doi: 10.1371/journal.pntd.0002921. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Carvalho R.G., Lourenço-de-Oliveira R., Braga I.A. Updating the Geographical Distribution and Frequency of Aedes albopictus in Brazil with Remarks Regarding Its Range in the Americas. Mem. Inst. Oswaldo Cruz. 2014;109:787–796. doi: 10.1590/0074-0276140304. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.