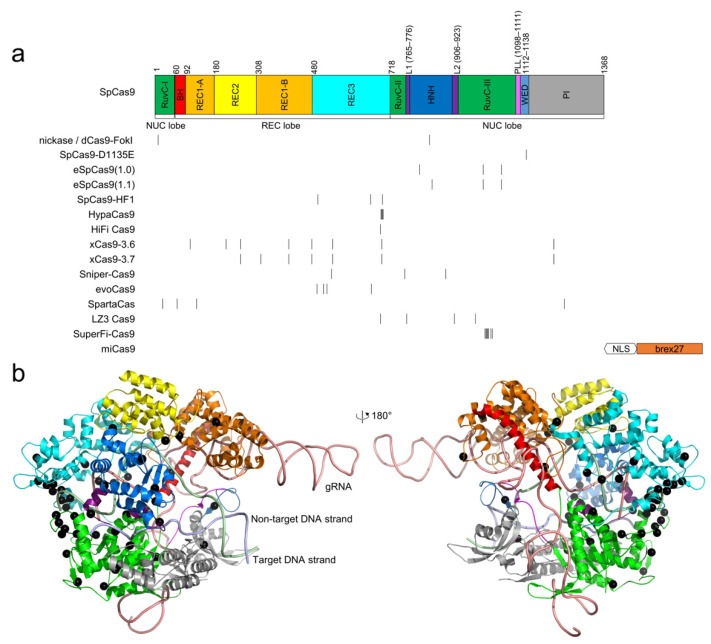
Figure 1.
The locations of high-fidelity SpCas9 mutations. (a) domain organization of SpCas9. Notably, the domain architecture varies in different literature; the architecture by Huai et al. [67] is used. The positions of the mutated residues for all high-fidelity variants are indicated by black vertical lines. miCas9 contains the fused brex27 motif connected by the SV40 NLS linker instead of point mutations. (b) the point mutations, shown as black spheres, are mapped into the structure of SpCas9 (PDB ID: 5F9R) [68]. The color scheme for SpCas9 is identical to that in (a).