Summary
Sensing carbohydrate availability is essential for plants to coordinate their growth and development. In Arabidopsis thaliana, TREHALOSE 6‐PHOSPHATE SYNTHASE 1 (TPS1) and its product, trehalose 6‐phosphate (T6P), are important for the metabolic control of development. tps1 mutants are embryo‐lethal and unable to flower when embryogenesis is rescued. T6P regulates development in part through inhibition of SUCROSE NON‐FERMENTING1 RELATED KINASE1 (SnRK1).
Here, we explored the role of SnRK1 in T6P‐mediated plant growth and development using a combination of a mutant suppressor screen and genetic, cellular and transcriptomic approaches.
We report nonsynonymous amino acid substitutions in the catalytic KIN10 and regulatory SNF4 subunits of SnRK1 that can restore both embryogenesis and flowering of tps1 mutant plants. The identified SNF4 point mutations disrupt the interaction with the catalytic subunit KIN10.
Contrary to the common view that the two A. thaliana SnRK1 catalytic subunits act redundantly, we found that loss‐of‐function mutations in KIN11 are unable to restore embryogenesis and flowering, highlighting the important role of KIN10 in T6P signalling.
Keywords: Arabidopsis thaliana, embryogenesis, flowering time, SnRK1 complex, TPS1, T6P pathway
Introduction
Flowering is an important process in the life cycle of plants and involves major physiological changes (Srikanth & Schmid, 2011; Romera‐Branchat et al., 2014; Song et al., 2015). Flowering time in Arabidopsis thaliana is under the control of several stimuli which are integrated in a complex genetic network that converges on floral integrator genes such as the florigen FLOWERING LOCUS T (FT) and SUPPRESSOR OF OVEREXPRESSION OF CO 1 (SOC1), which in turn control the expression of floral meristem identity genes such as LEAFY (LFY) and APETALA1 (AP1). Once activated, flowering commences with the induction of floral meristems at the flank of the shoot meristem, followed by internode elongation, or bolting.
Daylength is among the most important factors affecting flowering time. In A. thaliana, flowering is accelerated in response to long days (LD) and is under the regulation of the circadian clock, which regulates CONSTANS (CO) expression during the day with CO in turn activating FT transcription (Srikanth & Schmid, 2011; Romera‐Branchat et al., 2014; Song et al., 2015). Plant age governs flowering mainly by the function of two microRNAs, miR156 and miR172 (Huijser & Schmid, 2011). As plants mature, miR156 levels decrease, resulting in an upregulation of SQUAMOSA PROMOTER BINDING‐LIKE (SPL) genes. SPL transcription factors promote flowering directly by activating the floral homeotic genes, and indirectly by inducing miR172, which represses APETALA2‐like (AP2) floral repressors.
Carbohydrate availability has also been implicated in the transition from vegetative to reproductive development. Of particular importance in this regard is the phospho‐disaccharide trehalose 6‐phosphate (T6P) (Cabib & Leloir, 1958). T6P plays a key role in signalling carbohydrate availability in plants, thereby regulating a large number of physiological and developmental responses (Eastmond et al., 2002; Schluepmann et al., 2003; van Dijken et al., 2004; Gomez et al., 2006, 2010; Lunn et al., 2006; Satoh‐Nagasawa et al., 2006; Wingler et al., 2012; Wahl et al., 2013; Ponnu et al., 2020). Furthermore, T6P has been implicated in the feedback regulation of sucrose concentrations by restricting sucrose synthesis and/or promoting sucrose consumption, forming a robust T6P–sucrose nexus (Lunn et al., 2014; Yadav et al., 2014).
In plants, T6P synthesis is catalysed by TREHALOSE PHOSPHATE SYNTHASE (TPS) proteins. In A. thaliana, TPS1 is the major active isoform (Vandesteene et al., 2010; Yang et al., 2012; Fichtner et al., 2020). Consistently, homozygous A. thaliana tps1 mutants (tps1‐2) display embryo‐lethality (Eastmond et al., 2002). However, when embryo lethality is bypassed by ectopically expressing dexamethasone (DEX)‐inducible TPS1 (GVG::TPS1) during seed set (van Dijken et al., 2004; Wahl et al., 2013), homozygous tps1‐2 plants that remain in the vegetative phase and fail to flower can be recovered. Thus, TPS1 catalytic activity is critical for the metabolic control of key plant developmental processes and transitions (Fichtner et al., 2020).
T6P regulates development in part through SnRK1. SnRK1 is a heterotrimeric kinase complex that acts as a sugar/energy sensor and is required for normal plant function and for plant responses to various stress conditions that affect energy homeostasis and thereby plant fitness and survival (Polge & Thomas, 2007; Baena‐Gonzalez & Sheen, 2008). SnRK1 is a structural and functional homologue of the low‐energy stress‐activated yeast SNF1 and animal AMP‐activated kinase (AMPK). The AMPK/SNF1/SnRK1 kinase complexes are typically composed of three different subunits: a catalytic α subunit (SnRK1α1/KIN10 or SnRK1α2/KIN11 in A. thaliana), a regulatory β subunit and a regulatory γ subunit (Hedbacker & Carlson, 2008; Ghillebert et al., 2011; Hardie et al., 2012; Broeckx et al., 2016). The catalytic subunits contain a highly conserved N‐terminal serine/threonine kinase domain, with an activation or T‐loop that requires phosphorylation for kinase activity, and a large C‐terminal regulatory domain for interaction with the other subunits (Estruch et al., 1992; Hawley et al., 1996; Baena‐Gonzalez et al., 2007). The T‐loop (Thr175 in KIN10) is phosphorylated by the upstream SnRK1 activating kinases, SnAK1/GRIK2 and SnAK2/GRIK1 (Kong & Hanley‐Bowdoin, 2002; Shen & Hanley‐Bowdoin, 2006; Shen et al., 2009; Crozet et al., 2010; Glab et al., 2017), but also shows significant autophosphorylation (Baena‐Gonzalez et al., 2007; Ramon et al., 2019). In A. thaliana, KIN10 is broadly expressed and believed to be responsible for most of the SnRK1 kinase activity (Jossier et al., 2009; Williams et al., 2014). The β‐subunits (KINβs) act as complex scaffolds but also control kinase activity, substrate specificity and localization (Hedbacker et al., 2004; Polge & Thomas, 2007; Ghillebert et al., 2011; Emanuelle et al., 2015; Ramon et al., 2019). In plants, a single hybrid βγ subunit (SNF4 in A. thaliana) acts as the complex γ subunit (Ramon et al., 2013). SNF4 is an essential gene as no homozygous loss‐of‐function mutants were obtained (Ramon et al., 2013; Gao et al., 2016). The βγ subunit consists of a conserved γ subunit domain with four cystathionine β‐synthase (CBS) motifs and a carbohydrate‐binding module (CBM), typically only found in the β subunits in nonplant species.
In AMPK and SNF1, the γ subunit acts as the energy‐sensing module, competitively binding AMP, ADP and ATP. However, in plants nucleotide charge does not have an important regulatory signal (Ramon et al., 2013; Emanuelle et al., 2015). Instead, SnRK1 is active by default and inhibited by high energy availability (Ramon et al., 2019). Sugars such as sucrose and glucose suppress SnRK1 activity (Baena‐Gonzalez et al., 2007) and this repressive effect can be attributed at least in part to T6P, which (as a proxy for high sugar availability) was identified as an allosteric inhibitor of SnRK1 (Zhang et al., 2009). More recently, T6P was suggested to directly bind to KIN10 and interfere with its binding and phosphorylation by the upstream kinases (Zhai et al., 2018). In response to activation (derepression) by low energy status (e.g. in extended darkness), SnRK1 phosphorylates a range of enzymes and transcription factors to reprogram metabolism and gene expression. Direct activation of C‐ and S1‐class bZIP transcription factor dimers, for example, induces the expression of genes such as DARK INDUCED6/ASPARAGINE SYNTHASE1 (DIN6/ASN1) and SENESCENCE5 (SEN5), which can be used to monitor SnRK1 activity (Baena‐Gonzalez et al., 2007; Delatte et al., 2011; Dietrich et al., 2011; Mair et al., 2015).
The role of the T6P pathway in flowering time control has been mainly associated with FT induction in leaves and the age pathway and miR156 expression in the shoot apical meristem (SAM) (Wahl et al., 2013). To identify mutations that rescue the nonflowering phenotype, tps1‐2 GVG::TPS1 seeds were mutagenized with ethyl methanesulfonate (EMS).
We expected to recover (at least) two categories of mutants from this screen: bypass mutations, which would restore flowering to tps1‐2 but were not necessarily involved in T6P signalling, and mutations that would interfere with T6P signalling downstream of TPS1. Here we describe the identification of several alleles with amino acid substitutions in the SnRK1 subunits KIN10 and SNF4 that can restore both flowering and embryogenesis in tps1‐2 plants. We found that flowering rescue requires both an early induction of FT in the leaves and a later decrease of miR156 and subsequent induction of SPLs in the SAM. All newly identified kin10 alleles have mutations in the catalytic domain C‐lobe. While the mutated G163 and G178 residues are located in or near the catalytic cleft (with the conserved T176 in the activation‐ or T‐loop), mutation of the R259 residue, which is more distant from the T‐loop in the primary protein sequence but in close spatial proximity (Broeckx et al., 2016; Jumper et al., 2021), might affect activity more indirectly. The single amino acid substitutions in SNF4 abolish or reduce interaction with KIN10, thereby also affecting SnRK1 complex function. Importantly, mutations in KIN11 were unable to rescue the tps1‐2 mutant. Our results demonstrate that loss of KIN10, but not KIN11, can restore flowering and embryogenesis in the tps1‐2 mutant, providing a clear genetic link between the T6P pathway and KIN10, and indicate that KIN10–SNF4 interaction is required for adequate SnRK1 activity in planta.
Materials and Methods
Plant materials and growth conditions
All plants are A. thaliana in the Col‐0 background. The tps1‐2, tps1‐2 GVG::TPS1, ft‐10 (GABI_290E08) and snrk1α1‐3 (GABI_579E09) mutants have been described (Eastmond et al., 2002; van Dijken et al., 2004; Yoo et al., 2007; Mair et al., 2015). kin11cr mutants were created by CRISPR/Cas9 using the pHSE401 binary vector (Xing et al., 2014) and gRNA1 and gRNA2 (Supporting Information Table S1; Fig. S1). Plants were grown on soil under wide photosynthetically active radiation (PAR) spectrum LED lights (110–130 μmol m−2 s−1; CLF Plant Climatics, Wertingen, Germany) under LD (16 h : 8 h, light : dark) or short day (SD, 8 h : 16 h, light : dark) conditions, 65% relative humidity and 23°C. For dark‐induced starvation, plants grown for 14 d in LD were exposed to an additional 12 h of darkness before harvesting under green light. Plants for the RNA‐sequencing (RNA‐seq) experiment were grown in different cabinets from the flowering time experiments, but with CLF wide PAR spectrum LED lights (110–130 μmol m−2 s−1) and the same conditions and settings. All newly characterized kin10 and snf4 mutations were confirmed by genotyping. See Methods S1 for details.
EMS mutagenesis of tps1‐2 GVG:TPS1 and identification of suppressor mutants
Around 15 000 tps1‐2 GVG:TPS1 seeds were stratified at 4°C for 3 d before being treated with 25 ml of 0.4% EMS (Sigma) as described (Weigel & Glazebrook, 2002). M1 plants were grown on soil and sprayed with 1 μM DEX (Sigma) solution containing 0.02% Tween‐20 (Sigma) at 2 d intervals from 10 d after sowing (DAS). M2 seeds were collected as 300 pools of 40–50 M1 plants. Approximately 500 M2 plants were grown from each M2 pool under LD at 23°C and screened for mutants that flowered without application of DEX. The phenotype and the homozygous state of the tps1‐2 transposon insertion were confirmed in the M3 generation by genotyping using primers 366, 367 and 368 (Table S1).
Mapping by sequencing
Mapping of EMS‐induced single nucleotide polymorphisms (SNPs) and statistical analyses were performed as previously described (Ossowski et al., 2008; Schneeberger et al., 2009). For details see Methods S2.
Flowering and bolting time measurement
Flowering and bolting time were measured by counting the number of days when inflorescences reach 1 cm (bolting) after sowing and the total number of leaves originating from the main shoot meristem, respectively. A minimum of 15 plants from different seed batches were used for each genotype per experiment. Error bars represent the standard deviation (SD) of mean values and letters the statistical differences among the genotypes based on ANOVA and Tukey’s HSD test.
RT‐qPCR and RNA‐seq data analyses
RNA for real‐time quantitative PCR (RT‐qPCR) was extracted using a RNeasy Plant Mini Kit (Qiagen) or TRIzol® Reagent (Invitrogen). cDNA was synthesised using the RevertAid First Strand cDNA Synthesis Kit (Thermo Scientific, Waltham, MA, USA) and qPCR was performed on a Bio‐Rad CFX96 instrument. Primers used for RT‐qPCR are listed in Table S1. For a detailed description of the RT‐qPCR analysis and RNA‐seq data preprocessing, differential expression and differential usage analyses see Methods S3, S4.
RNA in situ hybridization
RNA in situ hybridization using a specific SOC1 antisense probe was carried out as previously described (Wahl et al., 2013; Olas et al., 2019). For details see Methods S5.
Vectors and cloning
For yeast‐two‐hybrid (Y2H) assays, the original pGADT7 and pGBKT7 vectors from Clontech/Invitrogen were modified to make them compatible with GreenGate (GG) cloning system ‘C’ modules (Lampropoulos et al., 2013). EcoR31I (or BsaI) sites were introduced into pGADT7 and pGBKT7 after the AD or BD, respectively, and a ccdB cassette and a chloramphenicol resistance gene were added between the EcoR31I sites. EcoR31I sites present in the backbone of pGADT7 and pGBKT7 were mutagenized by site‐directed mutagenesis. The final newly designed GreenGate‐compatible BD and AD vectors designated as pVZ022‐AD and pVZ023‐BD were verified by sequencing. PCR‐amplified and gel‐purified products were introduced into pVZ022‐AD and pVZ023‐BD by GreenGate cloning (Lampropoulos et al., 2013). Mutant variants of SNF4 were generated by site‐directed mutagenesis of SNF4‐AD and verified by sequencing. Phusion high‐fidelity DNA polymerase (Thermo Scientific) was used for all PCRs.
For transient expression assays, the KIN10, KIN11 and SNF4 coding sequences (CDSs) without the stop codon were amplified by PCR from A. thaliana Col‐0 cDNA and inserted into the HBT95 vector, in‐frame with a double haemagglutinin (HA) or a FLAG tag, yellow fluorescent protein (split‐YFP) or enhanced green fluorescent protein (GFP) tag (Sheen, 1996). The DIN6/At3g47340 and SEN5/AT3G15450 promoter–LUC reporter systems were previously described (Baena‐Gonzalez et al., 2007). To obtain the KIN10 and SNF4 mutant alleles, plasmid site‐directed mutagenesis was performed. To clone the KIN11 truncated protein coding sequences, we used primers kin11A and kin11cr2B or kin11cr3B. All constructs were confirmed by sequencing. All primers used for cloning are listed in Table S1.
Transient expression in leaf mesophyll protoplasts
Col‐0 plants were grown under a 12 h : 12 h, light : dark diurnal cycle with 75 μE cool white fluorescent light (F17T8/TL741/ALTO; Philips, Eindhoven, the Netherlands) for 4 wk at 21°C. Leaf mesophyll protoplast isolation and transfection were performed as described by Yoo et al. (2007). After PEG‐Ca2+‐mediated transfection, protoplasts were incubated under dim light (10 μE) for 6 h for LUC and GUS activity assays and immunoblot analyses or 16 h for GFP localization and bimolecular fluorescence complementation (BiFC) assays. For statistical analysis, one‐way ANOVA and Tukey’s HSD test were applied.
Promoter‐LUC activity assays
For LUC and GUS activity assays, 104 protoplasts were transfected with 10 μg DNA (CsCl gradient‐purified), as described in Ramon et al. (2019).
Subcellular localization studies
To observe the subcellular localization of the wild‐type and mutant KIN10 proteins, 4 × 104 protoplasts were transfected with a total of 30 μg GFP‐construct plasmid DNA (CsCl gradient‐purified) and 10 μg SC35‐like splicing factor 30 (SCF30)‐RFP (red fluorescent protein) nuclear DNA marker (CsCl gradient‐purified) and incubated for 16 h. GFP and RFP were visualized using confocal laser scanning microscopy (FV1000; Olympus Europe, Hamburg, Germany) with a 40× (oil) objective.
Bimolecular fluorescence complementation assays
To determine in vivo interactions, 4 × 104 protoplasts were transfected with a total of 30 μg split‐YFP construct plasmid DNA (CsCl gradient‐purified) and 10 μg SCF30‐RFP nuclear DNA marker (CsCl gradient‐purified) and incubated for 16 h. YFP and RFP were visualized using confocal laser scanning microscopy (FV1000; Olympus) with a 40× (1.3 oil) objective.
Immunoblot analysis
For detection of transiently expressed proteins, protoplasts were transfected with 20 μg DNA (CsCl gradient‐purified) and incubated for 6 h, after which 20 μl of loading buffer was added to the protoplast pellet and boiled for 5 min at 95°C. Immunoblotting was performed as described in Ramon et al. (2019).
Yeast‐two‐hybrid assays
All final AD and BD Y2H constructs were transformed into Y187 and AH109 yeast cells, respectively (Gietz & Schiestl, 2007). Y187 and AH109 cells with the introduced constructs were selected on single synthetic dropout (SD) media without leucine (−L) or tryptophan (−W), respectively. To test the protein binary interactions, the yeast mating system was used according to the Yeast protocols handbook (PT3024‐1‐Clontech 2001, Palo Alto, CA, USA). Protein–protein interactions were demonstrated by the activation of both HIS and β‐galactosidase reporters.
Results
Mutations in KIN10 and SNF4 subunits restore flowering in the tps1‐2 mutant
To identify genes that restore flowering and seed set in the nonflowering tps1‐2 GVG::TPS1 mutant, we carried out an EMS suppressor screen. In total, 106 putative mutants, which suppressed the nonflowering tps1‐2 GVG:TPS1 phenotype and produced seeds, were recovered in the M2 generation.
To identify the causal mutation in one of the suppressor lines, 160‐1, the mutant was backcrossed once to tps1‐2 GVG::TPS1. In total, 180 individual F2 segregant plants, corresponding to c. 25% of the population, showed the suppressor phenotype (BC1F2) without the application of DEX, and were used for mapping by sequencing. SNP analysis revealed a strong enrichment of EMS‐induced SNPs at the top of chromosome 3 in the BC1F2 population of line 160‐1 (Fig. 1a). One of the candidate genes containing a nonsynonymous SNP encodes the SnRK1 catalytic subunit KIN10 (At3g01090). As SnRK1 has previously been implicated in T6P signalling (Zhang et al., 2009; Schluepmann et al., 2012; Nunes et al., 2013), we considered this mutation as the prime candidate for restoring flowering in the tps1‐2 GVG::TPS1 suppressor line 160‐1.
Fig. 1.
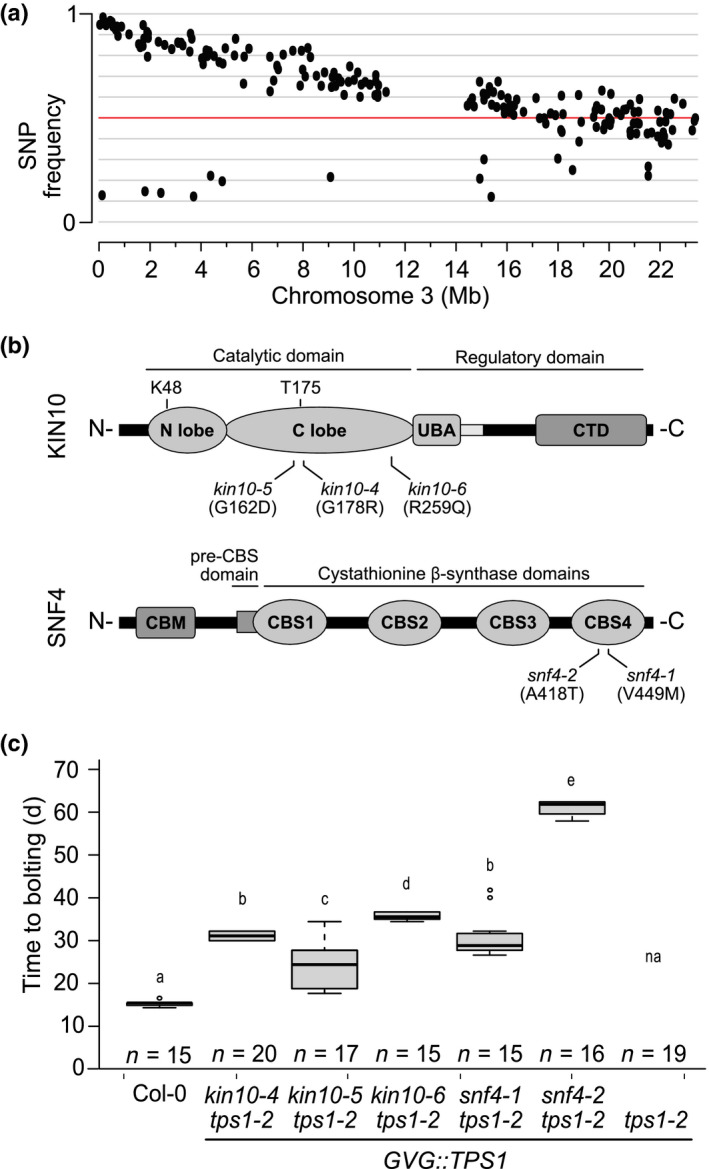
Mutations in KIN10 and SNF4 subunits restore flowering in the Arabidopsis tps1‐2 GVG::TPS1 mutant. (a) Frequency of ethyl methanesulfonate (EMS)‐induced single nucleotide polymorphisms (SNPs) on chromosome 3 in the BC2F2 population of 160‐1 plants. Red line indicates 50% SNP frequency. (b) Schematic representation of the location of the suppressor mutations on the KIN10 and SNF4 proteins. Top: KIN10 protein consisting of a catalytic domain including an N‐terminal N‐lobe and C‐terminal C‐lobe, which contain the conserved K48 residue important for phosphotransfer and the T‐loop with indicated conserved threonine (T175) residue; a ubiquitin‐associated (UBA) domain and linker sequence; and a C‐terminal domain (CTD), essential for the complex and other protein–protein interactions. Bottom: the SNF4 protein consisting of an N‐terminal carbohydrate‐binding module (CBM), a pre‐CBS domain, and four cystathionine β‐synthase (CBS) domains required for nucleotide binding and complex interactions. Black lines indicate the point mutations identified in this work and the resulting amino acid substitutions in parentheses. (c) Flowering time of EMS suppressor mutants. Plants were grown in LD conditions without dexamethasone application. A one‐way ANOVA Tukey’s test was applied and letters represent the statistical differences among genotypes (P < 0.001), and error bars represent SD; n, number of individuals; na, not applicable because plants do not flower.
To screen for additional kin10 alleles, we sequenced the genome of 64 additional suppressor mutants that displayed stable suppression of the tps1‐2 phenotype in the M3 generation (Table S2). After correcting for SNPs that were detected in multiple suppressor lines, 33 513 informative unique SNPs were identified and mapped to the genome (Table S3). From these data we identified three additional suppressor lines (170‐1, 199‐6, 232‐2‐1) that carried nonsynonymous amino acid substitutions in KIN10 (Table S4). Complementation crosses were carried out between three of the potential kin10 lines, 160‐1, 199‐6 and 232‐2‐1. F1 plants flowered without DEX application, indicating that the three tested suppressor mutants form one complementation group and confirming that the mutations in kin10 are causal for floral induction in the suppressor lines (Fig. S2). We refer to these new EMS‐induced alleles as kin10‐4 (160‐1), kin10‐5 (232‐2‐1) and kin10‐6 (199‐6) (Fig. 1b).
Closer examination of the SNP data led to the identification of four potentially deleterious mutations in the SNF4 subunit: three nonsynonymous amino acid substitutions and one potential splice site change (Tables S3, S4). Complementation crosses between two of these potential snf4 alleles, lines 125‐6‐1 and 154‐1‐1, flowered in the F1 generation without DEX application, suggesting strongly that the mutations in SNF4 were causal (Fig. S2). We refer to these new EMS‐induced snf4 mutant alleles as snf4‐1 and snf4‐2, respectively (Fig. 1b).
All kin10 or snf4 mutants that were confirmed by complementation assays were backcrossed to tps1‐2 GVG::TPS1 at least once and homozygous suppressor mutant lines were established. Flowering was restored to a similar extent in all the backcrossed lines, except for snf4‐2 tps1‐2 GVG::TPS1, which flowered significantly later than the other suppressor lines (Fig. 1c), indicating that snf4‐2 might be a hypomorphic SNF4 allele.
Due to the presumed functional redundancy of KIN10 and KIN11 (Baena‐Gonzalez et al., 2007; Jeong et al., 2015), we investigated whether the loss of a functional KIN11 subunit also rescues tps1‐2 flowering ability. For this, we created two CRISPR/Cas9 mutant KIN11 alleles (kin11cr) (Fig. S1) and crossed them into both tps1‐2 and tps1‐2 GVG::TPS1 plants. However, we were unable to recover a double kin11cr tps1‐2 mutant and kin11cr tps1‐2 GVG::TPS1 plants stalled in the vegetative phase, indicating that kin11cr mutations neither rescued embryo lethality nor flowering. kin11cr plants were indistinguishable from the wild‐type, as already reported for a kin11 T‐DNA knockout line (Jeong et al., 2015). To confirm that the mutant KIN11cr alleles do not produce any residual KIN11 activity, we transiently expressed the resulting truncated proteins, lacking a major part of the catalytic domain, in leaf mesophyll protoplasts. As expected, they no longer activate the DIN6‐LUC reporter with the kin11cr2 allele not even producing detectable protein levels, possibly due to nonsense‐mediated decay of the transcript and/or instability of the truncated protein (Fig. S1). To summarize, we found that mutations in KIN10 and SNF4, but not in KIN11, can restore flowering in the otherwise nonflowering tps1‐2 GVG::TPS1 background.
Mutations in KIN10 and SNF4 suppress tps1‐2 embryo lethality
Embryo development in homozygous tps1‐2 plants is arrested at the torpedo stage (Eastmond et al., 2002). Interestingly, embryogenesis is rescued in all the suppressor mutants, including the hypomorphic snf4‐2 tps1‐2 GVG::TPS1 mutant. However, these latter mutants produced only very few viable seeds (at most 10 per plant) compared to the other mutants. To exclude the possibility that the observed rescue of flowering and embryogenesis in the suppressor mutants was not caused by inadvertent activation of the GVG::TPS1 transgene, we crossed two of the kin10 alleles and one of the snf4 alleles with tps1‐2 heterozygous plants lacking GVG::TPS1. Homozygous mutations in the two SnRK1 subunits were able to restore flowering and embryo development in the homozygous tps1‐2 background (kin10‐4 tps1‐2; kin10‐5 tps1‐2; snf4‐1 tps1‐2), even in the absence of the GVG::TPS1 transgene (Fig. S3c,d). Moreover, the introduction of a previously characterized kin10 T‐DNA insertion, snrk1α1‐3 (Mair et al., 2015), into the tps1‐2 mutant restored flowering to a similar extent as the kin10 alleles recovered from the EMS suppressor screen (Fig. S4). Together, these findings confirm that mutations in both KIN10 and SNF4 can restore embryo development of the tps1‐2 mutant.
Importantly, similar to snrk1α1‐3 (Mair et al., 2015), all kin10 and snf4 alleles recovered from the EMS suppressor screen were phenotypically indistinguishable from wild‐type when introduced into the Col‐0 background, indicating that under the conditions tested, single loss‐of‐function mutations in SnRK1 subunits can only display a phenotype in a sensitized background such as tps1‐2.
Induction of flowering of the tps1‐2 mutant by SnRK1 mutations involves both the photoperiod‐dependent and age pathways
We observed that apart from the hypomorphic snf4‐2 tps1‐2 GVG::TPS1, all kin10 or snf4 tps1‐2 GVG::TPS1 double mutants are bolting at 34–38 DAS (Fig. 1c). To determine the time of flowering, we also measured the expression of AP1, a flower meristem identity gene, in apices of kin10‐5 tps1‐2 GVG::TPS1 and snf4‐1 tps1‐2 GVG::TPS1 plants. AP1 expression was detectable in wild‐type plants starting from 18 DAS and peaked at 22 DAS. In tps1‐2 GVG::TPS1 plants, no induction of AP1 could be detected. In both double mutants, AP1 became detectable only from 22 DAS, and further increased at 34 DAS (Fig. 2c). As expected, FT was highly expressed in Col‐0 leaves at 14 and 18 DAS (Fig. 2b), preceding AP1 expression (Fig. 2c). Surprisingly, even though FT expression increased transiently in the double mutants starting at 14 DAS, expression never reached wild‐type levels and was very low by the time AP1 expression became detectable (Fig. 2b,c). Transient FT expression in the suppressor mutants can be explained by upstream CO expression (Fig. 2a), which is at wild‐type levels at 14 DAS but drops to close to tps1‐2 GVG:TPS1 levels at later time points. This suggests that although the CO–FT module is initially activated in the suppressor mutants, this activation is not maintained and might not be sufficient to complete the transition to flowering. Indeed, we found that the SnRK1 suppressor mutations were capable of inducing flowering in the ft‐10 mutant background (Fig. 2f). Triple mutants flowered later than either ft‐10 or double kin10‐2 and snf4‐1 mutants (Fig. 2f), showing an additive effect. Expression of the FT paralogue TWIN SISTER OF FT (TSF) was also not significantly restored in the suppressor mutants except for at one time point (Fig. S5). These data suggest that the photoperiod‐dependent pathway is involved in initiating the floral transition in the double mutants, but that completion of flowering and bolting may require additional factors.
Fig. 2.
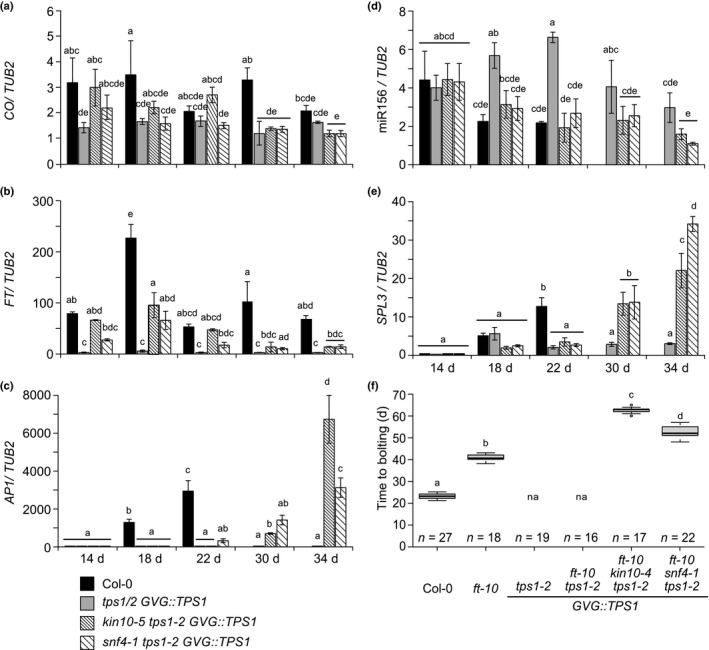
Regulation of the age and the photoperiod‐dependent pathways is important for SnRK1 to restore flowering in the Arabidopsis tps1 mutant. (a, b) CO (a) and FT (b) expression in leaves (whole rosettes) of 14‐ to 34‐d‐old plants in long day (LD) conditions. (c, d) Expression of AP1 (c), miR156 (d) and SPL3 (e) in apices of 14‐ to 34‐d‐old plants in LD conditions. ANOVA Tukey’s multiple comparisons test. Letters a–e represent statistical differences among genotypes and time points. (f) Flowering time of kin10‐5 tps1‐2 GVG::TPS1 and snf4‐1 tps1‐2 GVG::TPS1 in ft‐10 background under LD conditions. Plants were grown without dexamethasone (DEX) application. ANOVA Tukey’s multiple comparisons test. Letters a–d represent the statistical differences among genotypes (P < 0.001) and error bars represent SD; n, number of individuals; na, not applicable because plants do not flower.
Likely candidates are miR156 and its targets, the SPL transcription factors, which were previously shown to be compromised in the SAM of tps1‐2 GVG::TPS1 plants (Wahl et al., 2013). In agreement with this, we observed a significantly reduced abundance of miR156 in the suppressor mutants compared to tps1‐2 GVG::TPS1 plants at the time of bolting, 30 and 34 DAS (Fig. 2d), whereas the miR156 target, SPL3, was strongly induced (Fig. 2e). Moreover, we observed that the suppressor mutants could flower even under noninductive SD conditions, where the photoperiod pathway is inactive (Fig. S3a,b). This suggests that induction of flowering in the tps1‐2 GVG::TPS1 background by mutations in SnRK1 involves the spatiotemporal activation of both the photoperiod‐dependent and the age pathways.
To investigate the effects of tps1‐2 and the suppressor mutations on gene expression in an unbiased way, we carried out a transcriptome analysis using RNA isolated from apices of 18‐, 26‐ and 34‐d‐old Col‐0, tps1‐2 GVG::TPS1, kin10‐5 tps1‐2 GVG::TPS1 and snf4‐1 tps1‐2 GVG::TPS1 plants. For Col‐0, we collected inflorescences at 26 and 34 DAS because plants had already bolted. Principal component analysis (PCA) indicated that the first component corresponds to developmental changes from a vegetative meristem (Fig. S6a; left), via a transition state, to the inflorescence meristem observed after bolting in 34‐d‐old Col‐0 plants (Fig. S6a; right). Expression of AP1 was detected at low levels in Col‐0 at 18 DAS while it was barely detectable in tps1‐2 GVG::TPS1 or the suppressor mutants at 18 DAS (Fig. S6b) but became detectable in 26‐d‐old suppressor mutants and increased further in 34‐d‐old plants (Fig. S6b), consistent with our RT‐qPCR data (Fig. 2c). Similar results were obtained for the B‐ and C‐class genes APETALA3 (AP3) (Fig. S6c), PISTILLATA (PI) (Fig. S6d) and AGAMOUS (AG) (Fig. S6e). The second component, which explains 12% of the variation in that data set, appears to correspond to the age of the plants (Fig. S6a). In this context, it is interesting to note that even though the tps1‐2 GVG::TPS1 mutant does not undergo floral transition, its transcriptome is not static and follows the two suppressor mutants in the PCA plot over time (Fig. S6a).
Because both suppressor mutants showed clear signs of floral transition at 26 DAS in our RNA‐seq experiment, further analyses focused on 18‐d‐old plants. Of the 2040 genes that were significantly differentially expressed between Col‐0 and tps1‐2 GVG::TPS1, 254 were also differentially expressed in kin10‐5 tps1‐2 GVG::TPS1 and snf4‐1 tps1‐2 GVG::TPS1 when compared to tps1‐2 GVG::TPS1 (Fig. S7; Table S5). Cluster analysis indicated that 175 of these genes were significantly more highly expressed in tps1‐2 GVG::TPS1 than the other three genotypes, whereas expression of 75 genes that were downregulated in tps1‐2 GVG::TPS1 was significantly restored in the suppressor mutants (Fig. S7; Table S5). Among the latter were SOC1, LFY and FUL, three important flowering time and flower meristem identity genes. Expression of these genes was strongly reduced in the tps1‐2 GVG::TPS1 mutant and significantly restored in both suppressor mutants at 18 DAS and further increased at 26 and 34 DAS (Fig. S6). Since SOC1 is an early marker of flowering, our RNA‐seq data suggest that the suppressor mutants have already initiated the transition to flowering at 18 DAS even though bolting occurs 16–20 d later. To test this hypothesis, we monitored the temporal–spatial expression of SOC1 in the SAM of plants that were transferred from SD to LD to induce flowering in a synchronized manner. RNA in situ hybridization detected transient induction of SOC1 in the centre of the SAM in Col‐0 at the time of floral transition, 3 and 5 d after the shift (Fig. S8). In line with our hypothesis, induction of SOC1 in the centre of the SAM was weaker but persisted longer in the snf4‐1 tps1‐2 GVG::TPS1 mutant before floral primordia became apparent 10 d after the shift. However, SOC1 expression was confined to the flanks and was not readily detectable in the centre of the SAM in the kin10‐5 tps1‐2 GVG::TPS1 mutant at any time point analysed. It would thus appear that mutations in snf4‐2 and kin10‐5, even though the proteins are part of the same SnRK1 complex, affect spatial expression of SOC1 differently.
Consistent with our RT‐qPCR data, the expression of several miR156 targets was also restored in the suppressor mutants (Fig. S9). Specifically, we found that expression of SPL3, SPL4, SPL5 and SPL15 was strongly increased at later time points (26 and 34 DAS). Notably, the expression of SPL9, which has previously been implicated in the regulation of phase transitions (Zhang et al., 2019), was also restored in both mutants (Fig. S9). Together, our results support the idea that the age pathway is required for completion of the floral transition and bolting in the suppressor mutants.
tps1‐2 suppressor mutations in KIN10 affect SnRK1 activity
The observation that single amino acid substitutions in KIN10 can bypass the developmental defects in the tps1‐2 GVG::TPS1 mutant led us to investigate the underlying molecular mechanism. To test whether these mutations are affecting SnRK1 activity, we performed luciferase reporter assays in A. thaliana leaf mesophyll protoplasts (Baena‐Gonzalez et al., 2007). The mutant versions of the KIN10 protein identified in kin10‐4 (G178R), kin10‐5 (G162D) and kin10‐6 (R259Q) were unable to activate the SEN5:LUC and DIN6:LUC reporters, similarly to the known inactive KIN10 variants with mutations in key residues of the catalytic domain (K48M) and the kinase T‐loop (T175A) (Fig. 3a,b), even though they are efficiently expressed (Fig. 3c) and appropriately localized to the cytoplasm and the nucleus (Fig. 3f).
Fig. 3.
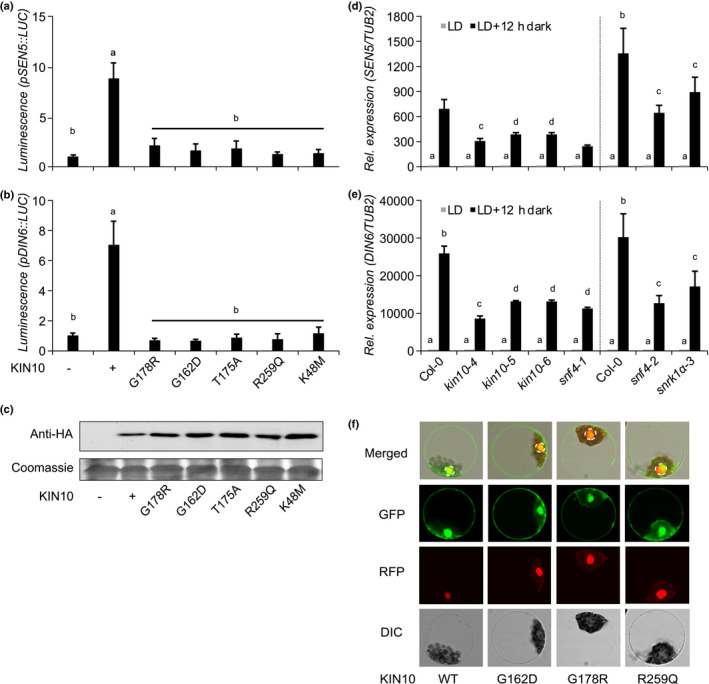
Mutations identified in Arabidopsis KIN10 and SNF4 result in nonfunctional KIN10 kinase. (a, b) SEN5 (a) and DIN6 (b) promoter activity in Arabidopsis thaliana leaf protoplasts upon transient expression of wild‐type and mutant KIN10 protein versions 6 h after transfection. G178R (kin10‐4), G162D (kin10‐5), R259Q (kin10‐6), K48M (KIN10 catalytic domain) and T175A (KIN10 T‐loop). Values are averages with standard deviations (n = 4). ANOVA Tukey’s multiple comparisons test was applied, letters represent the statistical differences (P < 0.001) and error bars represent SD. (c) Protein expression was assessed by immunoblot analysis with anti‐HA antibodies. Coomassie staining of Rubisco small subunit (RBCS) served as a loading control. (d, e) SEN5 (d) and DIN6 (e) endogenous expression in 14‐d‐old single kin10 and snf4 mutants. LD, long days. Vertical dashed black lines separate the RT‐qPCR results of two independent experiments. Error bars represent the SD of three biological replicates. ANOVA Tukey’s multiple comparisons test was applied and letters represent the statistical differences among genotypes (P < 0.001). (f) Subcellular localization of GFP‐tagged KIN10 proteins in A. thaliana leaf mesophyll protoplasts. An SCF30‐RFP nuclear marker was coexpressed. Dashed circles indicate the nucleus. DIC, differential interference contrast.
To further verify that the mutants are impaired in SnRK1 function in planta, we analysed expression of the SnRK1 target genes SEN5 and DIN6 in response to carbon starvation as a consequence of an artificially extended night. Our results show that the SEN5 and DIN6 induction, observed in Col‐0 plants, was attenuated in our kin10 and snf4 mutants, to a similar extent as in the previously published kin10 T‐DNA insertion line, snrk1α‐3 (Mair et al., 2015) (Fig. 3d,e). Together, these results indicate that the newly identified mutations, located in the catalytic domain, affect KIN10 activity, whereas the subcellular localization of the mutant proteins appeared to be unaffected when transiently expressed in protoplasts.
tps1‐2 suppressor mutations in SNF4 affect the interaction with KIN10
To the best of our knowledge, no viable snf4 mutations have been reported. In this study, we identified two mutant snf4 alleles based on their ability to rescue the nonflowering phenotype and suppress embryo lethality of the tps1‐2 GVG::TPS1 mutant. Interestingly, these snf4 mutations also resulted in reduced SnRK1 signalling activity (Fig. 3d,e). Since SNF4 is a noncatalytic subunit of the SnRK1 complex and the two mutations we identified are both located in the CBS4 domain (Fig. 1b), possibly involved in facilitating complex formation (Gissot et al., 2006; Ramon et al., 2013), we used Y2H and BiFC analysis to test whether the mutations affected the ability of SNF4 to interact with KIN10.
We detected a strong interaction between wild‐type SNF4 and KIN10 (Fig. 4a) in yeast, confirming previous results (Kleinow et al., 2000). The interaction was also detectable between KIN10 and the SNF4 variant encoded by the hypomorphic snf4‐2 allele (SNF4‐A418T), although the interaction was substantially weaker (Fig. 4a). By contrast, the protein encoded by the strong snf4‐1 allele (SNF4‐V449M) was unable to interact with KIN10 (Fig. 4a). These findings are corroborated by results from BiFC assays, which demonstrated that the mutant SNF4 proteins no longer effectively interact with KIN10 in the cytosol or nucleus of leaf cells (Figs 4b, S10) even though they were stably expressed (Fig. 4c). Upon extended exposure, only some small, isolated foci were observed in addition to chloroplast autofluorescence (Figs 4b, S10).
Fig. 4.
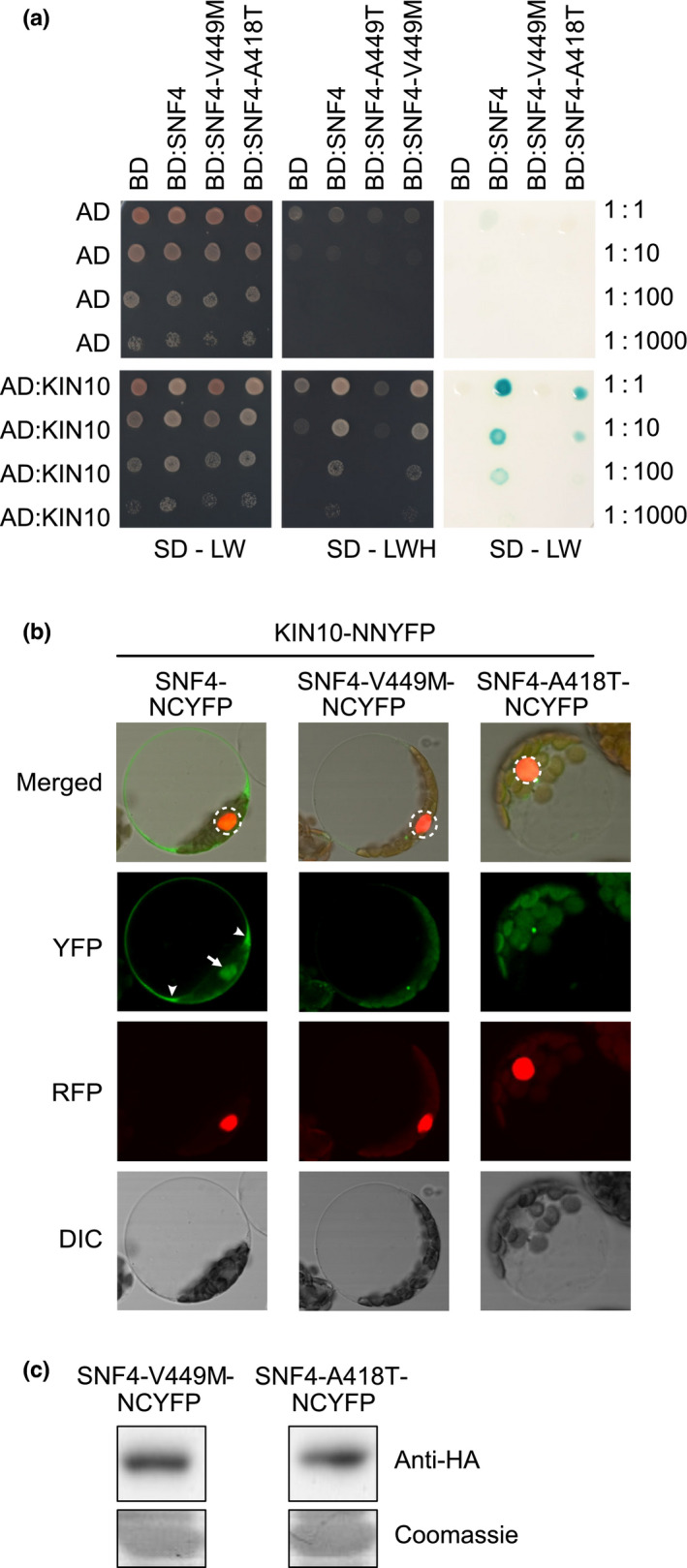
SNF4 protein mutant versions no longer participate in SnRK1 heterotrimeric complexes. (a) Arabidopsis SNF4 protein mutant versions (SNF4‐V449M and SNF4‐A418T) do not interact or interact weakly with KIN10 in a yeast two‐hybrid assay. Yeast colonies were grown in parallel in double dropout media (SD‐LW) (left panel) and triple dropout media (SD‐LWH) (middle panel) to demonstrate the presence of both bait and prey plasmids (SD‐LW) and protein–protein interactions with successful activation of the HIS reporter gene and growth in the absence of histidine (SD‐LWH). Right panel: interactions among bait and prey proteins were also confirmed based on expression of the β‐galactosidase enzyme, as indicated by the blue colour produced during enzymatic hydrolysis of the X‐gal substrate. Three serial dilutions were performed (1 : 10, 1 : 100, 1 : 1000) to show the strength of the interaction. (b) Bimolecular fluorescence complementation (BiFC) assay of the interaction between KIN10 and SNF4, SNF4‐V449M or SNF4‐A418T upon transient coexpression of the indicated HA‐tagged split‐YFP constructs, 16 h after transfection. An SCF30‐RFP nuclear marker was coexpressed. Dashed circles indicate the nucleus. White arrow and arrowheads mark nuclear and cytoplasmic YFP signals in wild‐type SNF4‐NCYFP, respectively. DIC, differential interference contrast. (c) Protein expression of the indicated HA‐tagged split‐YFP constructs was assessed by immunoblot analysis with anti‐HA antibodies. Coomassie staining of Rubisco small subunit (RBCS) served as a loading control. NCYFP, N‐terminal C‐YFP tag.
It was recently shown that the KINβ2 subunit can sequester KIN10 in the cytoplasm by myristoylation‐dependent membrane association, thereby inhibiting SnRK1 target gene activation. However, SNF4 can antagonize this inhibitory effect, possibly by competing interactions with the catalytic β subunit (Ramon et al., 2019). To test whether loss of the KIN10 interaction with the SNF4 mutant variants (SNF4‐A418T and SNF4‐V449M) attenuates SnRK1 target gene activation, we coexpressed the SNF4 wild‐type and mutant proteins with KIN10 and KINβ2 in A. thaliana leaf mesophyll protoplasts and monitored DIN6:LUC reporter activation. We found that coexpression of wild‐type SNF4 indeed relieved KINβ2 repression of KIN10 DIN6:LUC reporter activation but that the mutant SNF4 proteins were no longer able to do so (Fig. 5). Together, our data suggest that loss of the KIN10–SNF4 interaction is responsible for reduced SnRK1 signalling.
Fig. 5.
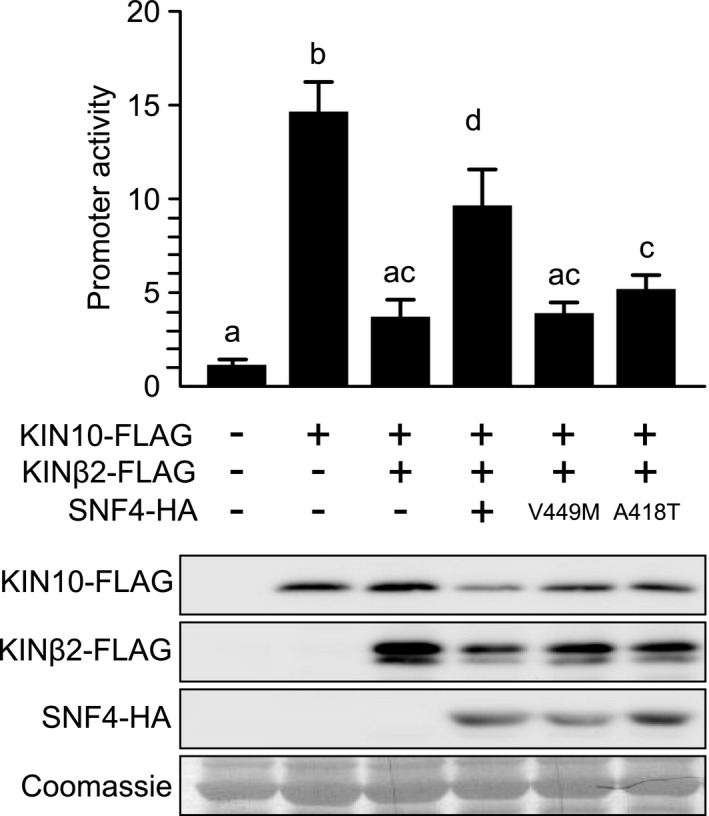
Mutated SNF4 versions no longer suppress KINβ2 inhibition of KIN10 activity. DIN6 promoter activity in Arabidopsis thaliana leaf mesophyll protoplasts upon transient expression of Arabidopsis KIN10, KINβ2 and SNF4 or SNF4‐V449M or SNF4‐A418T 6 h after transfection. Values are averages with standard deviations (n = 4). ANOVA Tukey’s multiple comparisons test was applied, and letters represent the statistical differences (P < 0.001). Protein expression was assessed by immunoblot analysis with anti‐HA and anti‐FLAG antibodies. RBSC served as a loading control.
Discussion
T6P and SnRK1 are central players in sensing carbohydrate availability and are essential antagonistic regulators of plant survival, growth and development, including embryogenesis and flowering (Eastmond et al., 2002; Schluepmann et al., 2003; van Dijken et al., 2004; Gomez et al., 2006; Baena‐Gonzalez et al., 2007; Wahl et al., 2013). Since it has been shown that KIN10 and TPS1 can modulate the circadian clock (Shin et al., 2017; Frank et al., 2018) and that overexpression of FT can induce flowering in a tps1 mutant (Wahl et al., 2013), we expected FT expression to be restored in the suppressor mutants. Surprisingly, whereas in wild‐type plants the induction of FT triggers floral formation within a few days (Fig. 2b), as indicated by AP1 expression (Fig. 2c), flowering occurred much later in the suppressor mutants despite an initial transient increase in FT expression (Fig. 2b,c). Based on the expression of flowering time and early flower development marker genes, such as SOC1 and LFY, it appears that the floral transition is only moderately delayed in the suppressor mutants (Fig. S6). This idea was confirmed by SOC1 RNA in situ hybridization (Fig. S8), which also revealed surprising differences in the spatial expression of SOC1 between the two suppressor mutants. How and why mutations in two subunits of the SnRK1 complex affect SOC1 patterning differently requires further investigation. Genetic analyses also showed that the suppressor mutations were sufficient to induce flowering in tps1‐2 GVG::TPS1 plants in the absence of a functional FT gene (Fig. 2f). Our findings indicate that floral transition is initiated but flower development and bolting are delayed in the suppressor mutants, suggesting the involvement of an additional mechanism.
Flower initiation and bolting are usually synchronized processes but can be uncoupled in early‐flowering A. thaliana accessions such as C24 and Ler‐1 (Miryeganeh et al., 2018). It has also been shown that plant age can induce flowering and bolting in A. thaliana (Huijser & Schmid, 2011) and that senescence and bolting are tightly linked processes (Hinckley & Brusslan, 2020). Interestingly, the SnRK1 complex, which promotes catabolic processes to ensure metabolic adaptation for increased cell viability and vitality, is known to affect senescence (Baena‐Gonzalez et al., 2007; Kim et al., 2017). For example, overexpression of KIN10 mimics cellular energy deprivation and delays natural leaf senescence (Baena‐Gonzalez et al., 2007; Kim et al., 2017). In addition, KIN10 directly interacts with and phosphorylates EIN3, thus delaying ethylene‐promoted organ senescence in plants (Kim et al., 2017). Since the floral transition appears to be extended in our suppressor mutants, and the miR156/SPL module, which forms the core of the age pathway, is misregulated in the SAM of the tps1‐2 GVG::TPS1 mutant, we hypothesized that the age pathway is involved in facilitating the timing and length of the floral transition and thus bolting in A. thaliana. The finding that at the time of bolting miR156 levels decrease and expression of several SPL genes increases at the SAM (Figs 2d,e, S9) supports this idea. In this context, note that constitutive overexpression of MIR156 has only a moderate effect on flowering time under LD but it causes extremely late flowering in SD (Schwab et al., 2005). Furthermore, tps1‐2 GVG::TPS1 mutants resemble SD‐grown plants in that the expression of FT and TSF is very much reduced even under LD conditions. This could explain why the moderate decrease of miR156 and the concomitant increase in SPLs have such a pronounced effect on flowering time and bolting in the suppressor mutants. This interpretation is also supported by the observation that the suppressor mutations can induce flowering in the ft‐10 mutant under LD conditions as well as in noninductive SD conditions (Fig. S3a,b).
Moreover, expression of MIR156a and MIR156c was reported to be repressed by sugars (Yang et al., 2013; Yu et al., 2013), suggesting regulation of the age pathway by the endogenous energy status. In A. thaliana, accumulation of sucrose and T6P specifically in LD conditions triggers FT expression in leaves to induce flowering (King et al., 2008), and at the time of the floral transition sucrose and T6P accumulate at the SAM (Madhusudanan & Nandakumar, 1983; Komarova & Milyaeva, 1991; Eriksson et al., 2006; Wahl et al., 2013). Thus, the requirement of both the age and the photoperiod pathway in regulating flowering and bolting downstream of the T6P pathway may not be so surprising. However, whereas SnRK1 might control the circadian clock and the photoperiod pathway quite directly (Shin et al., 2017; Frank et al., 2018), regulation by the age pathway might be more indirect. Interestingly, a recent study demonstrated that many bolting‐associated genes are also expressed during leaf senescence, suggesting that bolting could stimulate senescence‐related signalling in mature leaves (Hinckley & Brusslan, 2020). Given that SnRK1 has a negative role in plant ageing and senescence (Baena‐Gonzalez et al., 2007; Kim et al., 2017), it is possible that the loss of a functional SnRK1 could trigger the age pathway when the plant ages and thus bolting and flowering completion.
Another interesting observation is that we recovered multiple mutant alleles in KIN10 from our suppressor screen, but none in KIN11. This is unexpected as the two catalytic subunits of SnRK1, KIN10 and KIN11, are often considered functionally redundant. This is based on the observations that single mutants do not display any obvious phenotypes (Jeong et al., 2015), but the double knockout is lethal, and knocking down the expression of both genes causes severe developmental defects (Baena‐Gonzalez et al., 2007). A possible explanation is that our genetic screen might not have been saturated or mutations in KIN11 were missed due to low sequencing coverage. However, a more likely explanation is that KIN10 and KIN11 are not fully redundant, a suggestion that is supported by the finding that introducing mutations in KIN11 in the tps1‐2 or tps1‐2 GVG::TPS1 background was not sufficient to restore embryogenesis or the ability to flower. Together with the observation that overexpression of KIN10 and KIN11 have opposing effects on flowering (Williams et al., 2014), our findings suggest strongly that the two genes have only partially overlapping functions.
In contrast to the other SnRK1 subunits, SNF4 is encoded by a single gene in A. thaliana (Lumbreras et al., 2001; Gissot et al., 2006) and is essential for plant survival (Ramon et al., 2013; Gao et al., 2016). This suggests that the snf4 mutants we have identified retain some function. It was recently demonstrated that an interaction with SNF4 is required for KIN10 nuclear activity in the presence of KINβ2, which can retain KIN10 in the cytoplasm through myristoylation‐mediated membrane association (Ramon et al., 2019). Interestingly, the point mutations we have identified in SNF4 are located in the last cystathionine β‐synthetase domain (CBS4) near the C‐terminus, which mediates protein–protein interactions (Fig. 1b). Since the complete loss of function is lethal and nucleotide‐binding may not be an important regulatory mechanism in the plant SnRK1 complex, we speculated that the mutated SNF4 proteins may no longer efficiently interact with KIN10. This would also explain why the snf4 point mutants in many aspects phenocopy kin10 mutants (Figs 1c, 3a–e). In line with this hypothesis, we observed that the SNF4 mutant proteins did not interact or interacted only weakly with KIN10 (Fig. 4), resulting in reduced nuclear target gene activation in the presence of KINβ2 (Fig. 5). These findings support the idea that the SNF4 interaction is important for KIN10 function and thus essential for SnRK1 energy signalling.
Rather than being activated by low energy stress, the plant SnRK1 kinase appears to be active by default and repressed in energy‐rich conditions (Ramon et al., 2019). Consistently, T6P, which functions as a signal for sucrose availability, was identified as an SnRK1 inhibitor (Zhang et al., 2009). A recent study reported that T6P controls SnRK1 activity by inhibiting the GRIK/SnAK‐mediated SnRK1 catalytic subunits’ T‐loop phosphorylation (Zhai et al., 2018). A prediction from these observations is that high T6P levels in the SAMs of fully developed and photosynthetically active plants and/or LD conditions should repress SnRK1 activity, thereby enabling the transition to flowering and seed set. The finding that suppressor mutations in the KIN10 and SNF4 subunits, which should prevent unrestricted SnRK1 activity, restore flowering and embryogenesis in the tps1 mutant is in line with this hypothesis. Our results therefore shed light on the connection and opposing roles of T6P and SnRK1 signalling in the metabolic control of plant growth and development, from embryogenesis to flowering.
Author contributions
VZ designed and performed most of the experiments; JP performed the EMS suppressor screen, the mapping of suppressor line 160‐1 and the low‐coverage sequencing of 64 suppressor mutants with help from TL; JH performed the bioinformatics and SNP analyses; NC and FR performed the cellular assays; MM‐L and VW performed the RNA in situ analysis; NS assisted with the Y2H construct preparation and MS conceived the project; VZ and MS wrote the article with input from all the authors.
Supporting information
Fig. S1 KIN11 CRISPR/Cas9 mutations.
Fig. S2 Flowering time of F1 plants from complementation crosses among kin10 and snf4 alleles.
Fig. S3 Flowering time of suppressor mutants in SD and double mutants in LD lacking the TPS1 inducible construct.
Fig. S4 The kin10 T‐DNA line (snrk1α‐3) rescues tps1‐2 flowering.
Fig. S5 Relative expression of TSF in whole rosettes of 14‐ to 34‐d‐old plants.
Fig. S6 RNA‐seq data from Col‐0, tps1‐2 GVG::TPS1, kin10‐5 tps1‐2 GVG::TPS1 and snf4‐1 tps1‐2 GVG::TPS1 apices.
Fig. S7 Analysis of significantly differentially expressed genes in 18‐d‐old plants.
Fig. S8 Expression of SOC1 in SAM detected by RNA in situ hybridization.
Fig. S9 Expression of SPL genes in apices obtained by RNA‐seq.
Fig. S10 SNF4 protein mutant versions are no longer participating in SnRK1 heterotrimeric complexes.
Methods S1 Genotyping of kin10 and snf4 mutations.
Methods S2 Mapping of EMS‐induced mutations by high‐throughput sequencing.
Methods S3 RT‐qPCR information.
Methods S4 RNA‐seq data analyses.
Methods S5 SOC1 RNA in situ hybridization.
Table S1 List of oligonucleotides used in this study.
Table S2 Number of identified SNPs in individual EMS suppressor lines.
Table S3 Unique SNPs identified in all sequenced EMS suppressor lines.
Table S4 EMS suppressor lines bearing nonsynonymous mutations in KIN10 and SNF4.
Table S5 Differentially expressed genes in (a) tps1‐2 GVG::TPS1 vs Col‐0, and (b) the suppressor mutants and tps1‐2 GVG::TPS1.
Please note: Wiley Blackwell are not responsible for the content or functionality of any Supporting Information supplied by the authors. Any queries (other than missing material) should be directed to the New Phytologist Central Office.
Acknowledgements
We thank Johannes Hanson for his critical reading and comments on the manuscript. We thank the UPSC bioinformatics facility (https://bioinfomatics.upsc.se) for technical support with regard to the RNA‐seq data preprocessing and analyses. We would also like to acknowledge support from Uppsala Multidisciplinary Center for Advanced Computational Science for access to the UPPMAX computational infrastructure. We acknowledge funding to the UPSC through grants from VINNOVA and The Knut and Alice Wallenberg Foundation. Work in the Wahl group is supported by the BMBF (031B0191), DFG grants within the SPP1530 (WA3639/1‐2, 2‐1) and the Max Planck Society. This work was supported by a grant from the Fund for Scientific Research – Flanders (grant FWO G011720N) to FR, a personal fellowship (FWO 11C8819N) to NC and grants from the Deutsche Forschungsgemeinschaft as part of the Priority Program SPP1530 (SCHM1560/8‐1, 8‐2) and from Vetenskapsrådet (2015‐04617) to MS.
Contributor Information
Filip Rolland, Email: filip.rolland@kuleuven.be.
Markus Schmid, Email: markus.schmid@umu.se.
Data availability
Mapping by sequencing and RNA‐seq data are available from ENA, accession nos. PRJEB37882 and PRJEB47979, respectively. Other data supporting the findings of this study are available from the corresponding author upon reasonable request.
References
- Baena‐Gonzalez E, Rolland F, Thevelein JM, Sheen J. 2007. A central integrator of transcription networks in plant stress and energy signalling. Nature 448: 938–942. [DOI] [PubMed] [Google Scholar]
- Baena‐Gonzalez E, Sheen J. 2008. Convergent energy and stress signaling. Trends in Plant Science 13: 474–482. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Broeckx T, Hulsmans S, Rolland F. 2016. The plant energy sensor: evolutionary conservation and divergence of SnRK1 structure, regulation, and function. Journal of Experimental Botany 67: 6215–6252. [DOI] [PubMed] [Google Scholar]
- Cabib E, Leloir LF. 1958. The biosynthesis of trehalose phosphate. Journal of Biological Chemistry 231: 259–275. [PubMed] [Google Scholar]
- Crozet P, Jammes F, Valot B, Ambard‐Bretteville F, Nessler S, Hodges M, Vidal J, Thomas M. 2010. Cross‐phosphorylation between Arabidopsis thaliana sucrose nonfermenting 1‐related protein kinase 1 (AtSnRK1) and its activating kinase (AtSnAK) determines their catalytic activities. Journal of Biological Chemistry 285: 12071–12077. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Delatte TL, Sedijani P, Kondou Y, Matsui M, de Jong GJ, Somsen GW, Wiese‐Klinkenberg A, Primavesi LF, Paul MJ, Schluepmann H. 2011. Growth arrest by trehalose‐6‐phosphate: an astonishing case of primary metabolite control over growth by way of the SnRK1 signaling pathway. Plant Physiology 157: 160–174. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dietrich K, Weltmeier F, Ehlert A, Weiste C, Stahl M, Harter K, Droge‐Laser W. 2011. Heterodimers of the Arabidopsis transcription factors bZIP1 and bZIP53 reprogram amino acid metabolism during low energy stress. Plant Cell 23: 381–395. [DOI] [PMC free article] [PubMed] [Google Scholar]
- van Dijken AJ, Schluepmann H, Smeekens SC. 2004. Arabidopsis trehalose‐6‐phosphate synthase 1 is essential for normal vegetative growth and transition to flowering. Plant Physiology 135: 969–977. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eastmond PJ, van Dijken AJ, Spielman M, Kerr A, Tissier AF, Dickinson HG, Jones JD, Smeekens SC, Graham IA. 2002. Trehalose‐6‐phosphate synthase 1, which catalyses the first step in trehalose synthesis, is essential for Arabidopsis embryo maturation. The Plant Journal 29: 225–235. [DOI] [PubMed] [Google Scholar]
- Emanuelle S, Hossain MI, Moller IE, Pedersen HL, van de Meene AM, Doblin MS, Koay A, Oakhill JS, Scott JW, Willats WG et al. 2015. SnRK1 from Arabidopsis thaliana is an atypical AMPK. The Plant Journal 82: 183–192. [DOI] [PubMed] [Google Scholar]
- Eriksson S, Bohlenius H, Moritz T, Nilsson O. 2006. GA(4) is the active gibberellin in the regulation of LEAFY transcription and Arabidopsis floral initiation. Plant Cell 18: 2172–2181. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Estruch F, Treitel MA, Yang X, Carlson M. 1992. N‐terminal mutations modulate yeast SNF1 protein kinase function. Genetics 132: 639–650. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fichtner F, Olas JJ, Feil R, Watanabe M, Krause U, Hoefgen R, Stitt M, Lunn JE. 2020. Functional features of TREHALOSE‐6‐PHOSPHATE SYNTHASE1, an essential enzyme in Arabidopsis . Plant Cell 32: 1949–1972. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Frank A, Matiolli CC, Viana AJC, Hearn TJ, Kusakina J, Belbin FE, Wells Newman D, Yochikawa A, Cano‐Ramirez DL, Chembath A et al. 2018. Circadian entrainment in Arabidopsis by the sugar‐responsive transcription factor bZIP63. Current Biology 28: 2597–2606 e2596. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gao XQ, Liu CZ, Li DD, Zhao TT, Li F, Jia XN, Zhao XY, Zhang XS. 2016. The Arabidopsis KINβγ subunit of the SnRK1 complex regulates pollen hydration on the stigma by mediating the level of reactive oxygen species in pollen. PLoS Genetics 12: e1006228. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ghillebert R, Swinnen E, Wen J, Vandesteene L, Ramon M, Norga K, Rolland F, Winderickx J. 2011. The AMPK/SNF1/SnRK1 fuel gauge and energy regulator: structure, function and regulation. FEBS Journal 278: 3978–3990. [DOI] [PubMed] [Google Scholar]
- Gietz RD, Schiestl RH. 2007. Frozen competent yeast cells that can be transformed with high efficiency using the LiAc/SS carrier DNA/PEG method. Nature Protocols 2: 1–4. [DOI] [PubMed] [Google Scholar]
- Gissot L, Polge C, Jossier M, Girin T, Bouly JP, Kreis M, Thomas M. 2006. AKINβγ contributes to SnRK1 heterotrimeric complexes and interacts with two proteins implicated in plant pathogen resistance through its KIS/GBD sequence. Plant Physiology 142: 931–944. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Glab N, Oury C, Guerinier T, Domenichini S, Crozet P, Thomas M, Vidal J, Hodges M. 2017. The impact of Arabidopsis thaliana SNF1‐related‐kinase 1 (SnRK1)‐activating kinase 1 (SnAK1) and SnAK2 on SnRK1 phosphorylation status: characterization of a SnAK double mutant. The Plant Journal 89: 1031–1041. [DOI] [PubMed] [Google Scholar]
- Gomez LD, Baud S, Gilday A, Li Y, Graham IA. 2006. Delayed embryo development in the ARABIDOPSIS TREHALOSE‐6‐PHOSPHATE SYNTHASE 1 mutant is associated with altered cell wall structure, decreased cell division and starch accumulation. The Plant Journal 46: 69–84. [DOI] [PubMed] [Google Scholar]
- Gomez LD, Gilday A, Feil R, Lunn JE, Graham IA. 2010. AtTPS1‐mediated trehalose 6‐phosphate synthesis is essential for embryogenic and vegetative growth and responsiveness to ABA in germinating seeds and stomatal guard cells. The Plant Journal 64: 1–13. [DOI] [PubMed] [Google Scholar]
- Hardie DG, Ross FA, Hawley SA. 2012. AMPK: a nutrient and energy sensor that maintains energy homeostasis. Nature Reviews Molecular Cell Biology 13: 251–262. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hawley SA, Davison M, Woods A, Davies SP, Beri RK, Carling D, Hardie DG. 1996. Characterization of the AMP‐activated protein kinase kinase from rat liver and identification of threonine 172 as the major site at which it phosphorylates AMP‐activated protein kinase. Journal of Biological Chemistry 271: 27879–27887. [DOI] [PubMed] [Google Scholar]
- Hedbacker K, Carlson M. 2008. SNF1/AMPK pathways in yeast. Frontiers in Bioscience 13: 2408–2420. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hedbacker K, Townley R, Carlson M. 2004. Cyclic AMP‐dependent protein kinase regulates the subcellular localization of Snf1‐Sip1 protein kinase. Molecular and Cellular Biology 24: 1836–1843. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hinckley WE, Brusslan JA. 2020. Gene expression changes occurring at bolting time are associated with leaf senescence in Arabidopsis . Plant Direct 4: e00279. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huijser P, Schmid M. 2011. The control of developmental phase transitions in plants. Development 138: 4117–4129. [DOI] [PubMed] [Google Scholar]
- Jeong EY, Seo PJ, Woo JC, Park CM. 2015. AKIN10 delays flowering by inactivating IDD8 transcription factor through protein phosphorylation in Arabidopsis . BMC Plant Biology 15: 110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jossier M, Bouly JP, Meimoun P, Arjmand A, Lessard P, Hawley S, Grahame Hardie D, Thomas M. 2009. SnRK1 (SNF1‐related kinase 1) has a central role in sugar and ABA signalling in Arabidopsis thaliana . The Plant Journal 59: 316–328. [DOI] [PubMed] [Google Scholar]
- Jumper J, Evans R, Pritzel A, Green T, Figurnov M, Ronneberger O, Tunyasuvunakool K, Bates R, Žídek A, Potapenko A et al. 2021. Highly accurate protein structure prediction with AlphaFold. Nature 596: 583–589. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim GD, Cho YH, Yoo SD. 2017. Regulatory functions of cellular energy sensor SNF1‐related kinase1 for leaf senescence delay through ETHYLENE‐INSENSITIVE3 repression. Scientific Reports 7: 3193. [DOI] [PMC free article] [PubMed] [Google Scholar]
- King RW, Hisamatsu T, Goldschmidt EE, Blundell C. 2008. The nature of floral signals in Arabidopsis. I. Photosynthesis and a far‐red photoresponse independently regulate flowering by increasing expression of FLOWERING LOCUS T (FT). Journal of Experimental Botany 59: 3811–3820. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kleinow T, Bhalerao R, Breuer F, Umeda M, Salchert K, Koncz C. 2000. Functional identification of an Arabidopsis Snf4 ortholog by screening for heterologous multicopy suppressors of snf4 deficiency in yeast. The Plant Journal 23: 115–122. [DOI] [PubMed] [Google Scholar]
- Komarova EN, Milyaeva EL. 1991. Changes in content and distribution of starch in stem apices of bicolored coneflower during the period of flowering evocation. Soviet Plant Physiology 38: 46–51. [Google Scholar]
- Kong LJ, Hanley‐Bowdoin L. 2002. A geminivirus replication protein interacts with a protein kinase and a motor protein that display different expression patterns during plant development and infection. Plant Cell 14: 1817–1832. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lampropoulos A, Sutikovic Z, Wenzl C, Maegele I, Lohmann JU, Forner J. 2013. GreenGate – a novel, versatile, and efficient cloning system for plant transgenesis. PLoS ONE 8: e83043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lumbreras V, Alba MM, Kleinow T, Koncz C, Pages M. 2001. Domain fusion between SNF1‐related kinase subunits during plant evolution. EMBO Reports 2: 55–60. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lunn JE, Delorge I, Figueroa CM, Van Dijck P, Stitt M. 2014. Trehalose metabolism in plants. The Plant Journal 79: 544–567. [DOI] [PubMed] [Google Scholar]
- Lunn JE, Feil R, Hendriks JH, Gibon Y, Morcuende R, Osuna D, Scheible WR, Carillo P, Hajirezaei MR, Stitt M. 2006. Sugar‐induced increases in trehalose 6‐phosphate are correlated with redox activation of ADPglucose pyrophosphorylase and higher rates of starch synthesis in Arabidopsis thaliana . Biochemical Journal 397: 139–148. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Madhusudanan KN, Nandakumar S. 1983. Carbohydrate changes in shoot tip and subtending leaves during ontogenetic development of pineapple. Zeitschrift Für Pflanzenphysiologie 110: 429–438. [Google Scholar]
- Mair A, Pedrotti L, Wurzinger B, Anrather D, Simeunovic A, Weiste C, Valerio C, Dietrich K, Kirchler T, Nägele T et al. 2015. SnRK1‐triggered switch of bZIP63 dimerization mediates the low‐energy response in plants. eLife 4: e05828. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miryeganeh M, Yamaguchi M, Kudoh H. 2018. Synchronisation of Arabidopsis flowering time and whole‐plant senescence in seasonal environments. Scientific Reports 8: 10282. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nunes C, O'Hara LE, Primavesi LF, Delatte TL, Schluepmann H, Somsen GW, Silva AB, Fevereiro PS, Wingler A, Paul MJ. 2013. The trehalose 6‐phosphate/SnRK1 signaling pathway primes growth recovery following relief of sink limitation. Plant Physiology 162: 1720–1732. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Olas JJ, Van Dingenen J, Abel C, Dzialo MA, Feil R, Krapp A, Schlereth A, Wahl V. 2019. Nitrate acts at the Arabidopsis thaliana shoot apical meristem to regulate flowering time. New Phytologist 223: 814–827. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ossowski S, Schneeberger K, Clark RM, Lanz C, Warthmann N, Weigel D. 2008. Sequencing of natural strains of Arabidopsis thaliana with short reads. Genome Research 18: 2024–2033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Polge C, Thomas M. 2007. SNF1/AMPK/SnRK1 kinases, global regulators at the heart of energy control? Trends in Plant Science 12: 20–28. [DOI] [PubMed] [Google Scholar]
- Ponnu J, Schlereth A, Zacharaki V, Dzialo MA, Abel C, Feil R, Schmid M, Wahl V. 2020. The trehalose 6‐phosphate pathway impacts vegetative phase change in Arabidopsis thaliana . The Plant Journal 104: 768–780. [DOI] [PubMed] [Google Scholar]
- Ramon M, Dang TVT, Broeckx T, Hulsmans S, Crepin N, Sheen J, Rolland F. 2019. Default activation and nuclear translocation of the plant cellular energy sensor SnRK1 regulate metabolic stress responses and development. Plant Cell 31: 1614–1632. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ramon M, Ruelens P, Li Y, Sheen J, Geuten K, Rolland F. 2013. The hybrid four‐CBS‐domain KINβγ subunit functions as the canonical gamma subunit of the plant energy sensor SnRK1. The Plant Journal 75: 11–25. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Romera‐Branchat M, Andres F, Coupland G. 2014. Flowering responses to seasonal cues: what's new? Current Opinion in Plant Biology 21: 120–127. [DOI] [PubMed] [Google Scholar]
- Satoh‐Nagasawa N, Nagasawa N, Malcomber S, Sakai H, Jackson D. 2006. A trehalose metabolic enzyme controls inflorescence architecture in maize. Nature 441: 227–230. [DOI] [PubMed] [Google Scholar]
- Schluepmann H, Berke L, Sanchez‐Perez GF. 2012. Metabolism control over growth: a case for trehalose‐6‐phosphate in plants. Journal of Experimental Botany 63: 3379–3390. [DOI] [PubMed] [Google Scholar]
- Schluepmann H, Pellny T, van Dijken A, Smeekens S, Paul M. 2003. Trehalose 6‐phosphate is indispensable for carbohydrate utilization and growth in Arabidopsis thaliana . Proceedings of the National Academy of Sciences, USA 100: 6849–6854. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schneeberger K, Ossowski S, Lanz C, Juul T, Petersen AH, Nielsen KL, Jorgensen JE, Weigel D, Andersen SU. 2009. SHOREmap: simultaneous mapping and mutation identification by deep sequencing. Nature Methods 6: 550–551. [DOI] [PubMed] [Google Scholar]
- Schwab R, Palatnik JF, Riester M, Schommer C, Schmid M, Weigel D. 2005. Specific effects of microRNAs on the plant transcriptome. Developmental Cell 8: 517–527. [DOI] [PubMed] [Google Scholar]
- Sheen J. 1996. Ca2+‐dependent protein kinases and stress signal transduction in plants. Science 274: 1900–1902. [DOI] [PubMed] [Google Scholar]
- Shen W, Hanley‐Bowdoin L. 2006. Geminivirus infection up‐regulates the expression of two Arabidopsis protein kinases related to yeast SNF1‐ and mammalian AMPK‐activating kinases. Plant Physiology 142: 1642–1655. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shen W, Reyes MI, Hanley‐Bowdoin L. 2009. Arabidopsis protein kinases GRIK1 and GRIK2 specifically activate SnRK1 by phosphorylating its activation loop. Plant Physiology 150: 996–1005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shin J, Sanchez‐Villarreal A, Davis AM, Du SX, Berendzen KW, Koncz C, Ding Z, Li C, Davis SJ. 2017. The metabolic sensor AKIN10 modulates the Arabidopsis circadian clock in a light‐dependent manner. Plant, Cell & Environment 40: 997–1008. [DOI] [PubMed] [Google Scholar]
- Song YH, Shim JS, Kinmonth‐Schultz HA, Imaizumi T. 2015. Photoperiodic flowering: time measurement mechanisms in leaves. Annual Review of Plant Biology 66: 441–464. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Srikanth A, Schmid M. 2011. Regulation of flowering time: all roads lead to Rome. Cellular and Molecular Life Sciences 68: 2013–2037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vandesteene L, Ramon M, Le Roy K, Van Dijck P, Rolland F. 2010. A single active trehalose‐6‐P synthase (TPS) and a family of putative regulatory TPS‐like proteins in Arabidopsis . Molecular Plant 3: 406–419. [DOI] [PubMed] [Google Scholar]
- Wahl V, Ponnu J, Schlereth A, Arrivault S, Langenecker T, Franke A, Feil R, Lunn JE, Stitt M, Schmid M. 2013. Regulation of flowering by trehalose‐6‐phosphate signaling in Arabidopsis thaliana . Science 339: 704–707. [DOI] [PubMed] [Google Scholar]
- Weigel D, Glazebrook J. 2002. Arabidopsis: a laboratory manual. Cold Spring Harbor, NY, USA: Cold Spring Harbor Laboratory Press. [Google Scholar]
- Williams SP, Rangarajan P, Donahue JL, Hess JE, Gillaspy GE. 2014. Regulation of sucrose non‐fermenting related kinase 1 genes in Arabidopsis thaliana . Frontiers in Plant Science 5: 324. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wingler A, Delatte TL, O'Hara LE, Primavesi LF, Jhurreea D, Paul MJ, Schluepmann H. 2012. Trehalose 6‐phosphate is required for the onset of leaf senescence associated with high carbon availability. Plant Physiology 158: 1241–1251. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xing HL, Dong L, Wang ZP, Zhang HY, Han CY, Liu B, Wang XC, Chen QJ. 2014. A CRISPR/Cas9 toolkit for multiplex genome editing in plants. BMC Plant Biology 14: 327. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yadav UP, Ivakov A, Feil R, Duan GY, Walther D, Giavalisco P, Piques M, Carillo P, Hubberten H‐M, Stitt M et al. 2014. The sucrose‐trehalose 6‐phosphate (Tre6P) nexus: specificity and mechanisms of sucrose signalling by Tre6P. Journal of Experimental Botany 65: 1051–1068. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang HL, Liu YJ, Wang CL, Zeng QY. 2012. Molecular evolution of trehalose‐6‐phosphate synthase (TPS) gene family in Populus, Arabidopsis and rice. PLoS ONE 7: e42438. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang L, Xu ML, Koo Y, He J, Poethig RS. 2013. Sugar promotes vegetative phase change in Arabidopsis thaliana by repressing the expression of MIR156A and MIR156C . eLife 2: e00260. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yoo SD, Cho YH, Sheen J. 2007. Arabidopsis mesophyll protoplasts: a versatile cell system for transient gene expression analysis. Nature Protocols 2: 1565–1572. [DOI] [PubMed] [Google Scholar]
- Yu S, Cao L, Zhou CM, Zhang TQ, Lian H, Sun Y, Wu JQ, Huang JR, Wang GD, Wang JW. 2013. Sugar is an endogenous cue for juvenile‐to‐adult phase transition in plants. eLife 2: e00269. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhai Z, Keereetaweep J, Liu H, Feil R, Lunn JE, Shanklin J. 2018. Trehalose 6‐Phosphate positively regulates fatty acid synthesis by stabilizing WRINKLED1. Plant Cell 30: 2616–2627. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang H, Zhang L, Han JY, Qian ZY, Zhou BY, Xu YM, Wu G. 2019. The nuclear localization signal is required for the function of squamosa promoter binding protein‐like gene 9 to promote vegetative phase change in Arabidopsis . Plant Molecular Biology 100: 571–578. [DOI] [PubMed] [Google Scholar]
- Zhang Y, Primavesi LF, Jhurreea D, Andralojc PJ, Mitchell RA, Powers SJ, Schluepmann H, Delatte T, Wingler A, Paul MJ. 2009. Inhibition of SNF1‐related protein kinase1 activity and regulation of metabolic pathways by trehalose‐6‐phosphate. Plant Physiology 149: 1860–1871. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Fig. S1 KIN11 CRISPR/Cas9 mutations.
Fig. S2 Flowering time of F1 plants from complementation crosses among kin10 and snf4 alleles.
Fig. S3 Flowering time of suppressor mutants in SD and double mutants in LD lacking the TPS1 inducible construct.
Fig. S4 The kin10 T‐DNA line (snrk1α‐3) rescues tps1‐2 flowering.
Fig. S5 Relative expression of TSF in whole rosettes of 14‐ to 34‐d‐old plants.
Fig. S6 RNA‐seq data from Col‐0, tps1‐2 GVG::TPS1, kin10‐5 tps1‐2 GVG::TPS1 and snf4‐1 tps1‐2 GVG::TPS1 apices.
Fig. S7 Analysis of significantly differentially expressed genes in 18‐d‐old plants.
Fig. S8 Expression of SOC1 in SAM detected by RNA in situ hybridization.
Fig. S9 Expression of SPL genes in apices obtained by RNA‐seq.
Fig. S10 SNF4 protein mutant versions are no longer participating in SnRK1 heterotrimeric complexes.
Methods S1 Genotyping of kin10 and snf4 mutations.
Methods S2 Mapping of EMS‐induced mutations by high‐throughput sequencing.
Methods S3 RT‐qPCR information.
Methods S4 RNA‐seq data analyses.
Methods S5 SOC1 RNA in situ hybridization.
Table S1 List of oligonucleotides used in this study.
Table S2 Number of identified SNPs in individual EMS suppressor lines.
Table S3 Unique SNPs identified in all sequenced EMS suppressor lines.
Table S4 EMS suppressor lines bearing nonsynonymous mutations in KIN10 and SNF4.
Table S5 Differentially expressed genes in (a) tps1‐2 GVG::TPS1 vs Col‐0, and (b) the suppressor mutants and tps1‐2 GVG::TPS1.
Please note: Wiley Blackwell are not responsible for the content or functionality of any Supporting Information supplied by the authors. Any queries (other than missing material) should be directed to the New Phytologist Central Office.
Data Availability Statement
Mapping by sequencing and RNA‐seq data are available from ENA, accession nos. PRJEB37882 and PRJEB47979, respectively. Other data supporting the findings of this study are available from the corresponding author upon reasonable request.


