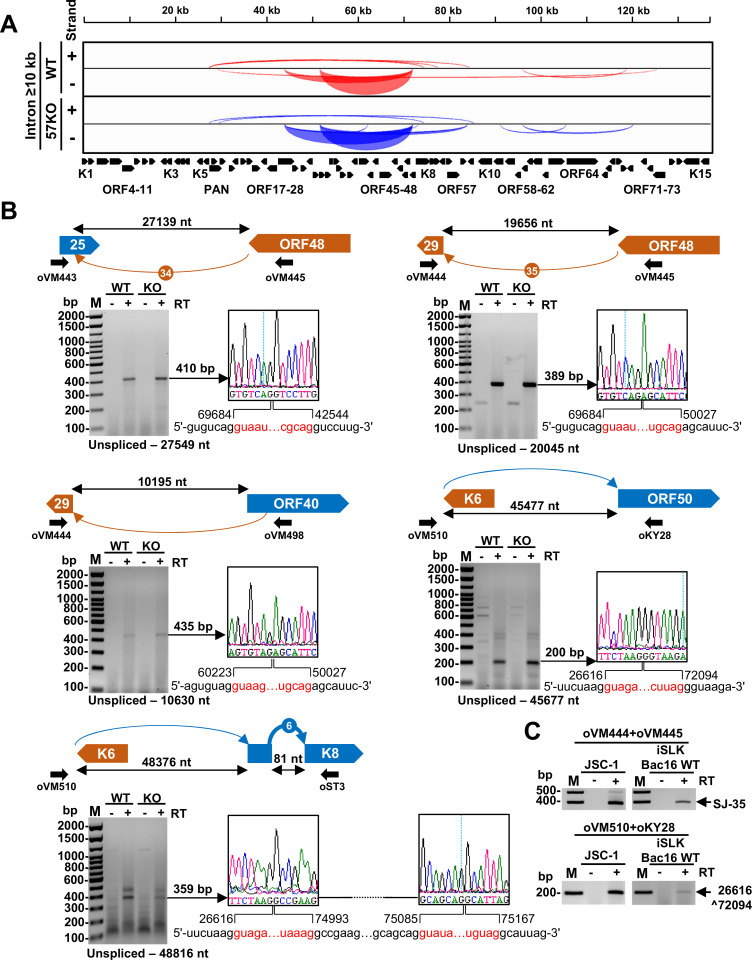
Fig 7. Long-range KSHV RNA splice identified in BCBL-1 cells with lytic KSHV infection.
(A) The Sashimi plot showing all splice junctions with an intron size ≥10 kb supported with at least 2 splice junction reads. See S2 Table for details. (B) Validations of selected long-range RNA splicing events by RT-PCR on total RNA from the WT genome or 57KO genome in the cells with lytic KSHV replication induced by VA for 24 h. Above the gel is a schematic showing a selected splicing event in orange arches for minus DNA strand-derived RNA transcripts and blue arches for plus DNA strand-derived RNA transcripts, relative genomic locations, predicted intron sizes, and primers used for RT-PCR. The arches lacking number-based nomenclature did not pass ≥10 of supporting splice reads (see S2 Table). Below is the gel electrograph of obtained RT-PCR products. The obtained RT-PCR products were reamplified and confirmed by Sanger sequencing. See sequencing primers in S6 Table. The chromatographs on the right show the identified splice junctions with the exonic sequences marked in black and the intronic sequences spliced out shown in red. The numbers represent exon-exon junction positions in the KSHV genome (GenBank Acc. No. HQ404500) after RNA splicing. (C) The conservation of long-range splicing events in JSC-1 and iSLK/Bac16 cells. Total RNA from the cells with induced KSHV lytic infection was used in RT-PCR using primers showed in (B). See S4A Fig for more details.