Figure 7. Effect of nerve growth factor silencing in cardiomyocytes on co‐cultured sympathetic neurons.
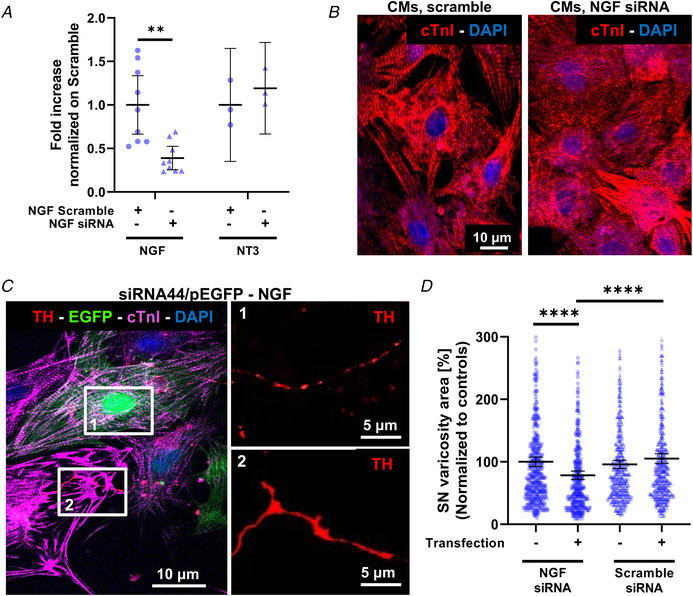
A , RTqPCR on extracts from cultured cardiomyocytes (CMs) transfected with scramble siRNA (white bars) or nerve growth factor (NGF) siRNA (black bars). Data distribution is represented by the individual values. Mean and error bars, representing 95% confidence intervals, are shown. Differences among groups were determined using an unpaired t test. (**, P < 0.01; n = 6 samples for each group in three independent cell preparations). B, confocal immunofluorescence (IF) of cultured CMs transfected with either scramble siRNA or NGF siRNA. Cells were stained with an antibody against cardiac troponin I (cTnI). Nuclei were counterstained with DAPI. C, confocal IF of 7‐day SN/CM co‐cultures in which CMs were co‐transfected with green fluorescent protein GFP and NGF siRNA. Cells were co‐stained with antibodies against tyrosine hydroxylase (TH) and cTnI. Nuclei were counterstained with DAPI. Images on the right are high magnifications of boxed areas on the left panel and show neuronal processes innervating, respectively, GFP+/NGF‐silenced (1) or control (2) CMs. D, quantification of the mean area of TH+ sites contacting un‐transfected, scramble‐siRNA transfected or NGF‐silenced CMs, in 7‐day co‐cultures maintained in the absence of exogenous NGF. Data distribution is represented by the individual values. Mean and error bars, representing 95% confidence intervals, are shown. Differences among groups were determined using the Mann–Whitney test. (** **, P < 0.0001; n > 400 neuronal processes for each condition, in three independent cell preparations). [Colour figure can be viewed at wileyonlinelibrary.com]
