Figure 1.
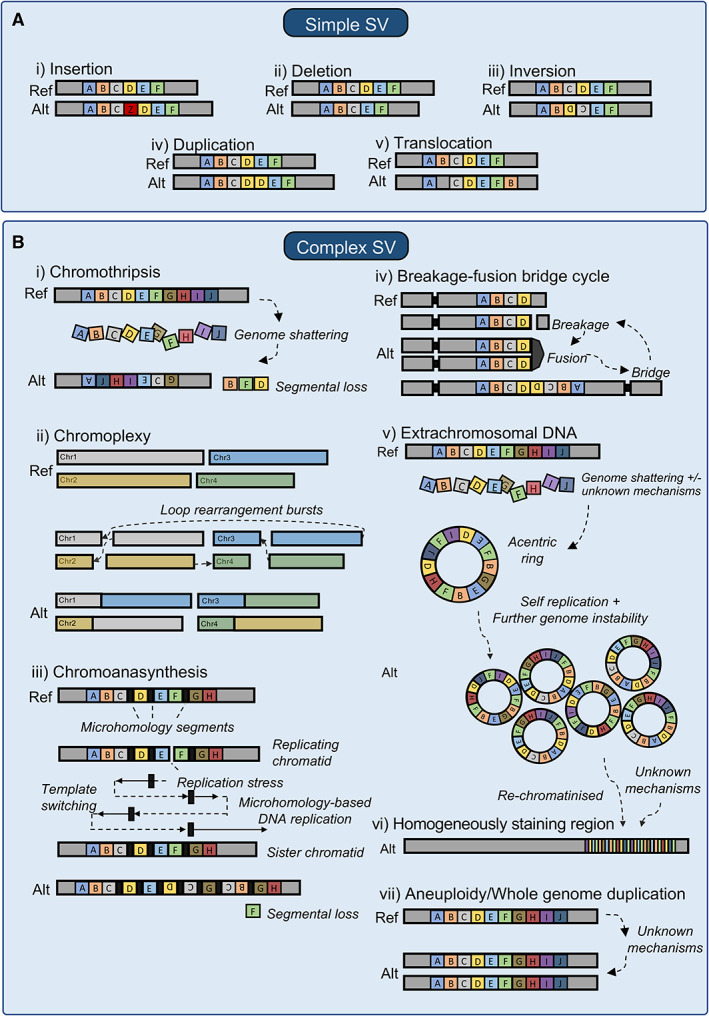
The diverse structural variant (SV) landscape in tumours. (A) Simple SVs include (i) insertions where segments of DNA are added, (ii) deletions where DNA segments are lost, (iii) inversions where DNA segments are in the opposite orientation, (iv) duplications where extra copies of DNA segments are present, and (v) translocations where a section of DNA is joined onto another section at a different genomic location. (B) Complex SVs include: (i) chromothripsis where the genome is shattered and the pieces joined back together at random, (ii) chromoplexy where interdependent translocations and deletions are joined together in a chain, (iii) chromoanasynthesis, which is a pattern of localised amplification and short stretches of breakpoint microhomology, (iv) breakage‐fusion‐bridge cycles, where telomeres are lost, dicentric chromosomes form before breaking due to mitotic stress, (v) extrachromosomal DNAs (ecDNAs), where large segments of DNA form acentric circles that often contain oncogenes and regulatory regions; these ecDNAs may be reincorporated into the chromosome, as (vi) homogeneously staining regions, and (vii) aneuploidy and whole‐genome duplication where some or all chromosomes have additional or double the number of copies, respectively.
