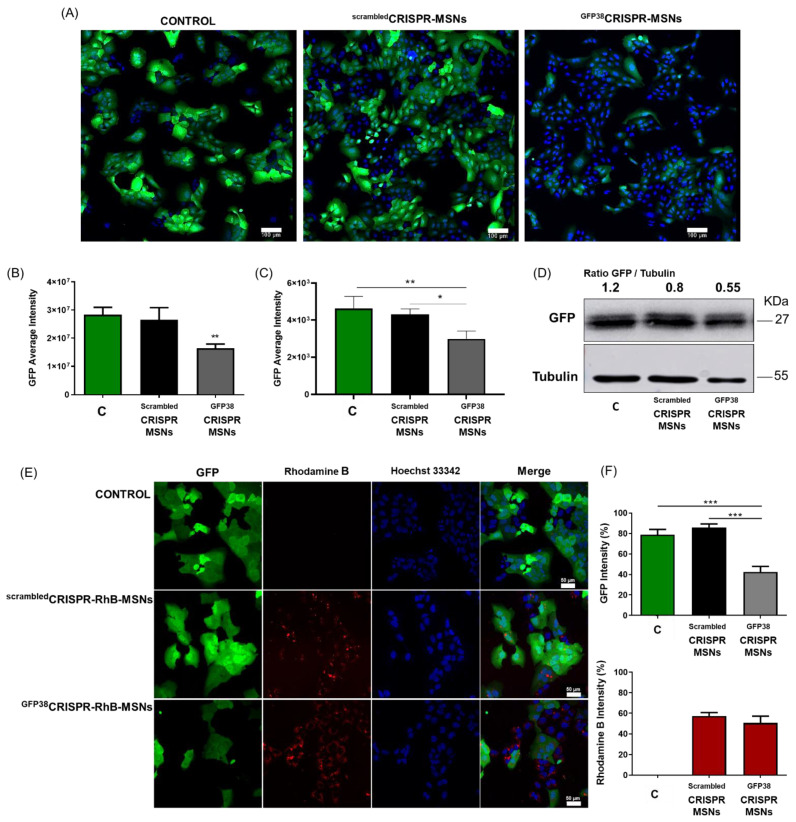
Figure 3.
Dual gene editing and cargo delivery in the U-2 OS-GFP cells. (A) Confocal microscopy images of genome editing by the CRISPR-Cas9 system delivered by MSNs as nanocarriers. Transfection efficiency is judged by fluorescent intensity and the proportion of cells in the population showing GFP expression. Green depicts the GFP cells and blue marks the nucleus with Hoechst 33342. The scale bar represents 10 µm. Representative images of the confocal experiments in at least three independent experiments. (B) GFP quantification by the confocal images analysis by using the Image J software. Data represent the mean ± SEM of at least three independent experiments. The control cells (untreated) are referred to as C. A statistical analysis was performed by applying a one-way ANOVA with multiple comparisons (* p < 0.05, ** p < 0.025, *** p < 0.001). (C) GFP quantification by IN Cell Analyzer 2200. Data represent the mean ± SEM of at least three independent experiments. A statistical analysis was performed by applying a one-way ANOVA with multiple comparisons (* p < 0.05, ** p < 0.025, *** p < 0.001). (D) Western blot analysis and quantification of the GFP levels expressed in the cell lysates of the CRISPR-MSNs editing studies. Representative images of at least three independent experiments. (E) Confocal microscopy images of genome editing and cargo delivery. Transfection efficiency is judged by fluorescent intensity and the proportion of cells in the population showing GFP (green) expression and delivery efficiency by the fluorescent intensity of Rhodamine B (red) and the nucleus in blue marked with Hoechst 33342. The scale bar represents 10 µm. Representative images of the confocal experiments in at least three independent experiments. (F) GFP (up) and Rhodamine B (down) quantification by the confocal images analysis by using the Image J software. The untreated cells, the negative control, are referred to as C. Data represent the mean ± SEM of at least three independent experiments. A statistical analysis was performed by applying a one-way ANOVA with multiple comparisons (* p < 0.05, ** p < 0.025, *** p < 0.001).