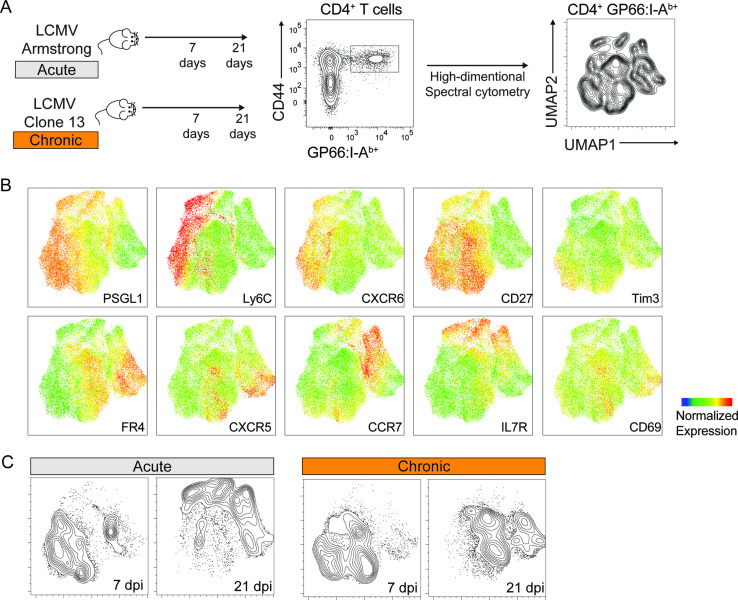
Figure 1. Phenotypic characterization of virus-specific CD4+ T cells by spectral flow cytometry.
Spleen GP66:I-Ab+ CD4+ T cells were analyzed 7 and 21 days after infection with LCMV Armstrong and Clone 13. (A) Schematic of experimental procedures. Uniform Manifold Approximation and Projection (UMAP) visualization was calculated based on the expression of 14 markers on virus-specific CD4+ T cells pooled from 5 animals. (B) Expression of selected markers shown on the UMAP as in (A). (C) CD4+ T cells from each condition were highlighted as contour lines on the UMAP. Experiment with 4–5 mice per group, representative of two independent experiments. See also Figure 1—figure supplement 1 for spectral cytometry analysis including GP33:H2Db+ CD8+ T cells, and for surface marker panels used to characterize T cell populations.