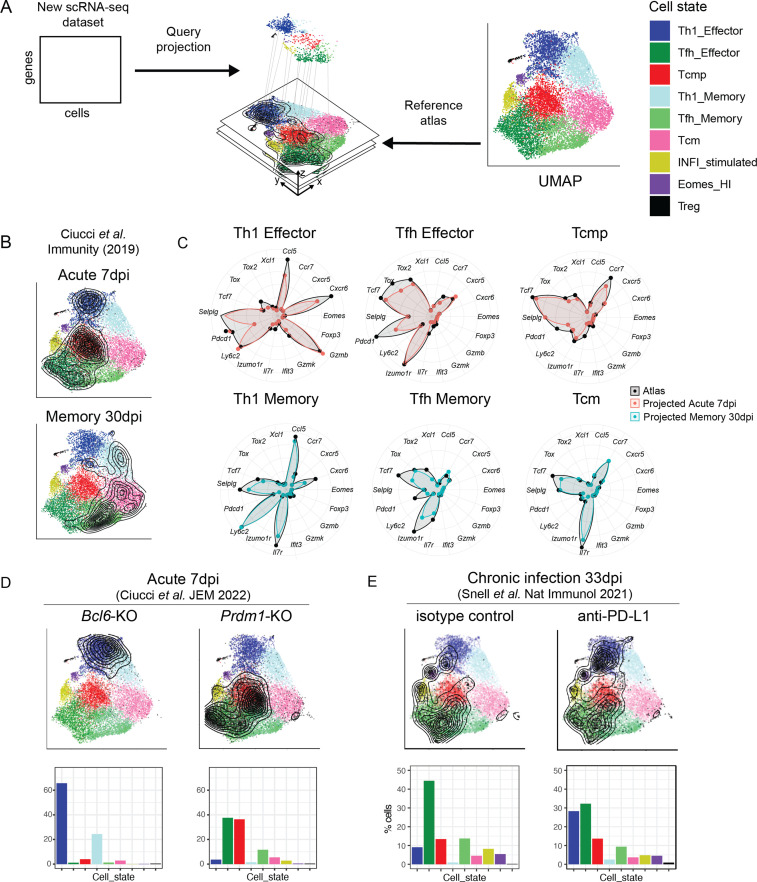
Figure 5. Interpretation of CD4+ T cell states in external datasets by projection into the reference map.
(A) Independent scRNA-seq datasets can be projected into the CD4+ T cell reference map using the ProjecTILs algorithm and interpreted in the context of the space and cell states of the reference. (B) UMAP embeddings for the external data by Ciucci et al., 2019 at 7 and 30 days after acute LCMV infection projected into the reference map. Contour lines indicate the density of projected cells. (C) Radar plots showing average expression profiles of a panel of CD4+ T cell marker genes, for the projected cells from Ciucci et al., 2019 (blue and red) compared to the reference map profiles (black) grouped by predicted subtype. (D) UMAP embeddings for projected scRNA-seq data of Bcl6- and Prdm1-deficient virus-specific CD4+ T cells isolated 7 days after acute infection with LCMV Armstrong (data from Ciucci et al., 2022). (E) UMAP embeddings for projected scRNA-seq data of virus-specific CD4+ T cells isolated 33 days after chronic infection with LCMV Clone13, in animals treated with anti-PDL1 or isotype control (data from Snell et al., 2021). Barplots in the bottom row indicate the percentage of cells projected in each reference subtype.