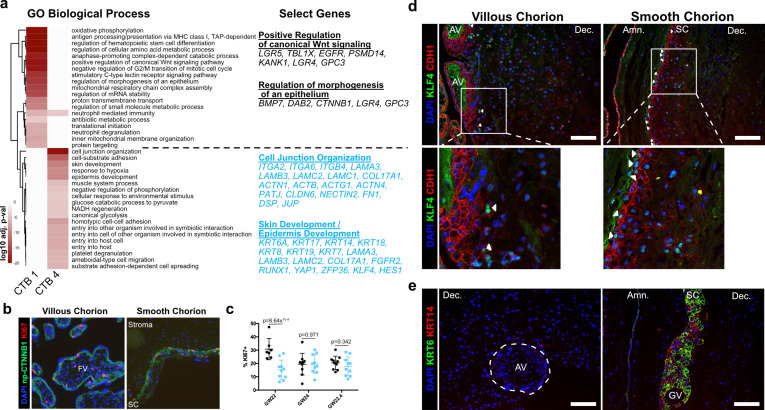
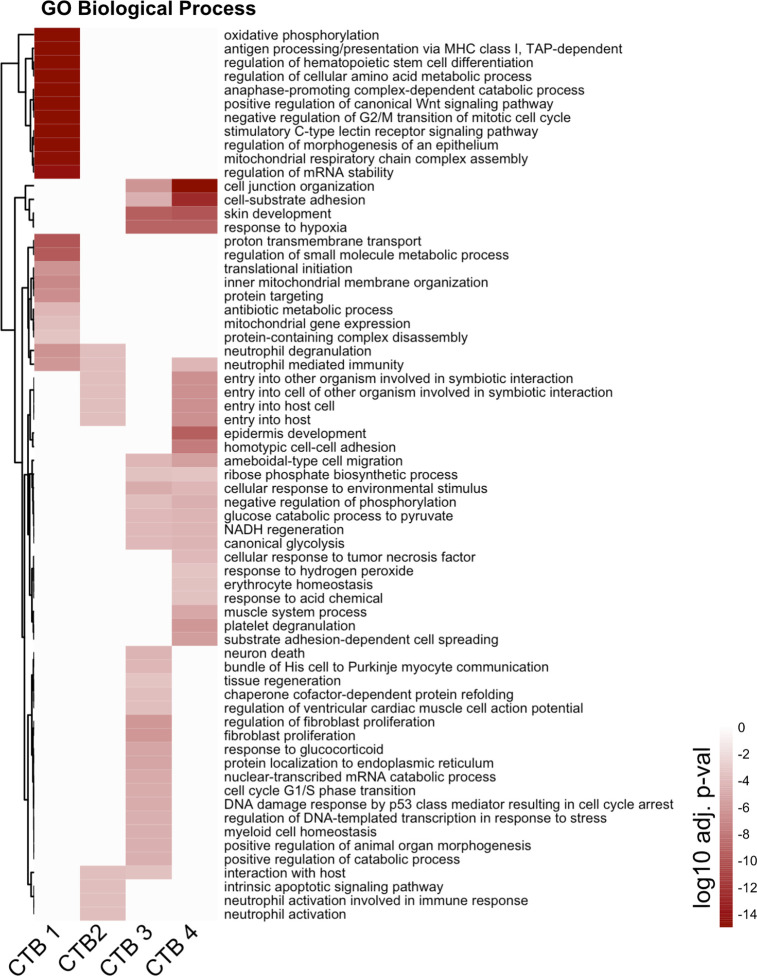
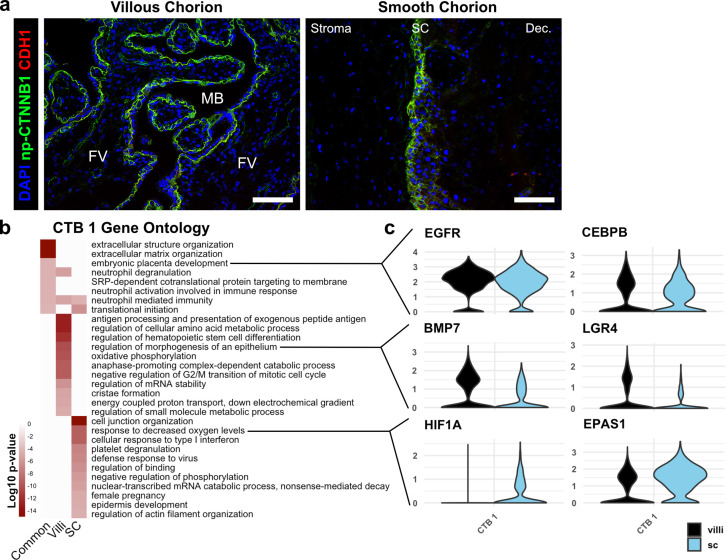
Figure 4. Smooth chorion-specific cytotrophoblasts (SC-CTBs) express a distinct epidermal transcriptional program.
(a) Heatmap of gene ontology analysis adjusted p-values. Dark red corresponds to the lowest p-values and white represents p-values greater than 0.0005. Ontology categories are organized by hierarchical clustering along the y-axis. Marker genes for each cluster were used as inputs for the analysis. A subset of genes in selected categories are listed at the right. Categories and genes enriched in CTB 1 or CTB 4 are in black or blue, respectively. (b) Representative immunofluorescence co-localization of non-phosphorylated CTNNB1 and KI67 in the VC (left) and SC (right). (c) Quantification of the percent of np-CTNNB1 cells with KI67 expression in each region in three placental samples. Each dot represents the percentage in one field of view (at least seven per region per sample) as an estimate of mitotic cells per population. Percentages for the villous chorion (VC) region are shown in black and the SC region in blue. (d) Immunofluorescence co-localization of CDH1 and KLF4 protein in the VC (left) or SC (right). Arrowheads denote CDH1+/KLF4+ cells. (e) Immunofluorescence co-localization of KRT14 and KRT6 protein in the VC (left) or SC (right). The outline of the anchoring villi (AV) is denoted by the white dashed line. For all images, nuclei were visualized by DAPI stain; scale bar = 100 μm. Abbreviations: FV = floating villi; SC = smooth chorion epithelium; Amn. = amnion; Dec. = decidua; GV = ghost villi.