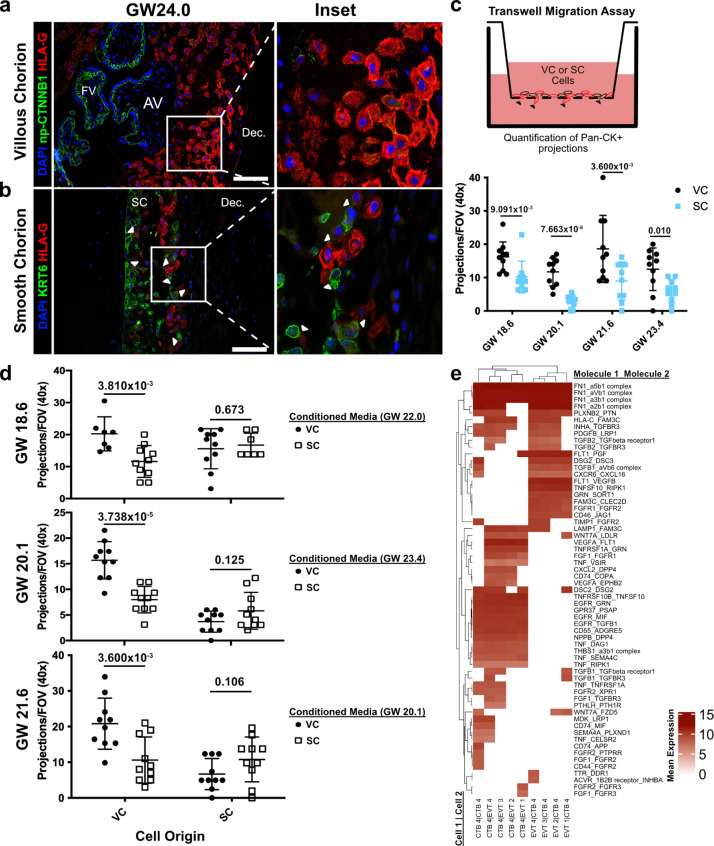
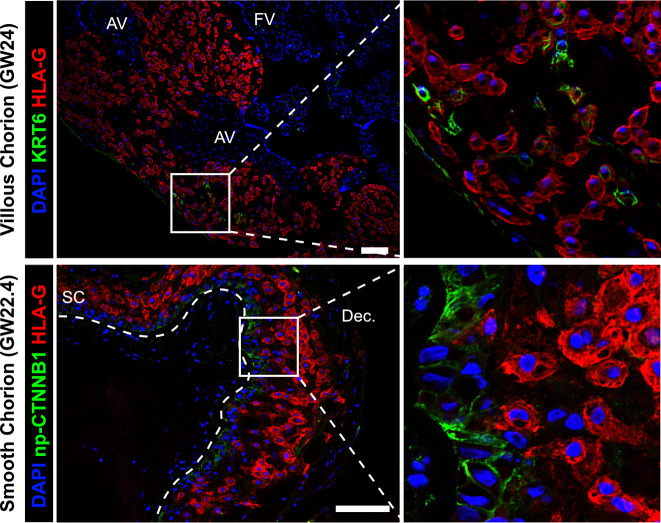
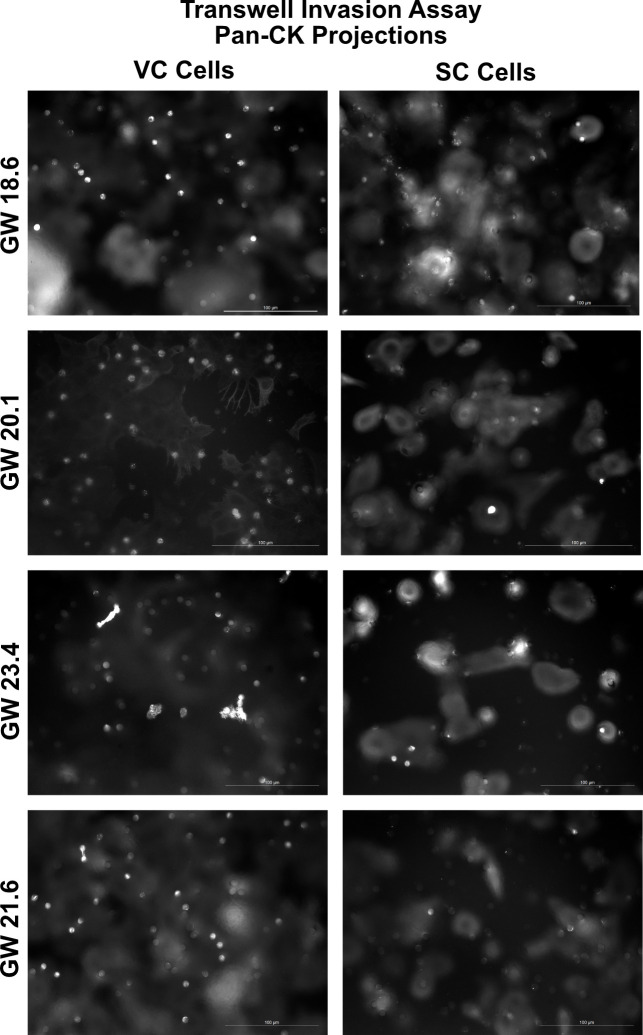
Figure 6. Cytotrophoblasts (CTBs) of the SC inhibit extravillous trophoblast (EVT) invasion.
(a) Immunofluorescence co-localization of np-CTNNB1 and HLA-G in the VC. (b) Immunofluorescence co-localization of KRT6 and HLA-G in the SC. Arrowheads denote CTB and EVT interactions. (c) Schematic of the transwell invasion assay (top). Cells from either chorionic region were plated in the upper chamber of the transwell. After 39 hr of culture, the transwell membrane was fixed and stained with a Pan-cytokeratin antibody. The projections through the membrane are denoted by black arrowheads and quantified below. Results from the VC derived cells are shown in black, and the results from the SC derived cells are shown in blue. (d) Results from three biological replicates from each chorionic region cultured with conditioned medium from either VC or SC cells. The gestational ages of the plated cells are shown adjacent to the y-axis. The gestational ages of the cells from which conditioned medium was collected are noted in the legends at the right. The results for cells cultured with VC cell conditioned medium are denoted by black dots, and the results for those cultured in SC cell conditioned medium are denoted by open squares. p-Values were determined by t-test and are listed above each comparison. (e) Predicted receptor-ligand interactions from CellPhoneDB. The strength of interaction is estimated by mean expression and is plotted in the heatmap. Receptor-ligand interactions and cell pairs are listed such that Molecule 1 is expressed by Cell 1 and Molecule 2 is expressed by Cell 2. For all images, nuclei were visualized by DAPI staining; scale bar = 100 μm. Abbreviations: AV = anchoring villi; FV = floating villi; SC = smooth chorion epithelium; Dec. = decidua.