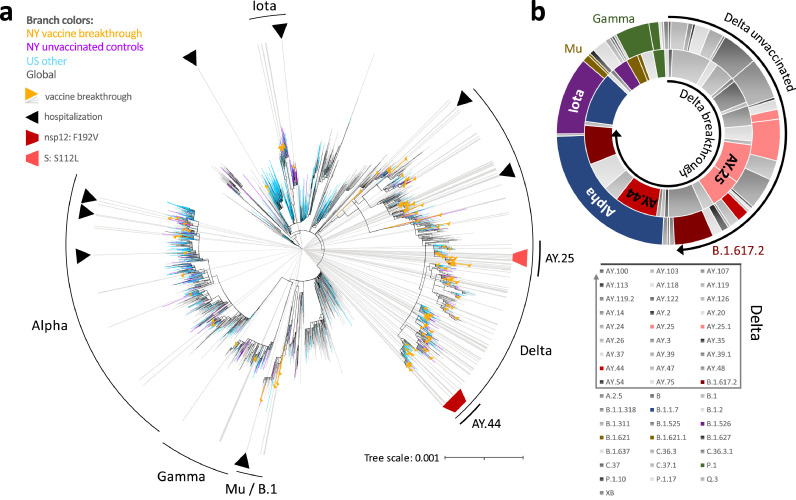
Figure 2.
Phylogenetic analysis and variant distribution of SARS-CoV-2 vaccine breakthrough and unvaccinated control sequences. a, Maximum likelihood (IQ) tree of 3511 SARS-CoV-2 full genome sequences (base pairs 202-29,666 according to Wuhan-Hu-1 as reference), including 132 vaccine breakthrough (orange) and 283 unvaccinated control SARS-CoV-2 sequences from the NYU Langone Health cohort (greater NYC area) (purple) together with 920 other US (non-NYU; cyan) and 2176 global (non-US; black) reference sequences. The substitution scale of the tree, generated with 1000 bootstrap replicates and Wuhan/WH01/2019-12-26 as root, is indicated at the bottom right. Vaccine breakthrough sequences are highlighted by orange triangles (as branch symbols) and grey rays radiating from the root to the outer rim of the tree. Hospitalizations among vaccine breakthrough infections are indicated by black triangles. The variants responsible for most vaccine breakthrough infections are labelled. The Delta plus spike:S112L (AY.25) and nsp12:F192V (AY.44) sub-lineages are labelled and highlighted with light and carmine red trapezoid symbols, respectively. b, Double-donut plot to compare the variant distribution of breakthrough (inner ring) and unvaccinated control sequences (outer ring). The most abundant variants and Delta subvariants (highlighted by black arrows) are shown in colour and labelled in the plot (outer ring only). All detected variants (Pango lineages) and their colour code in the plot are shown below.