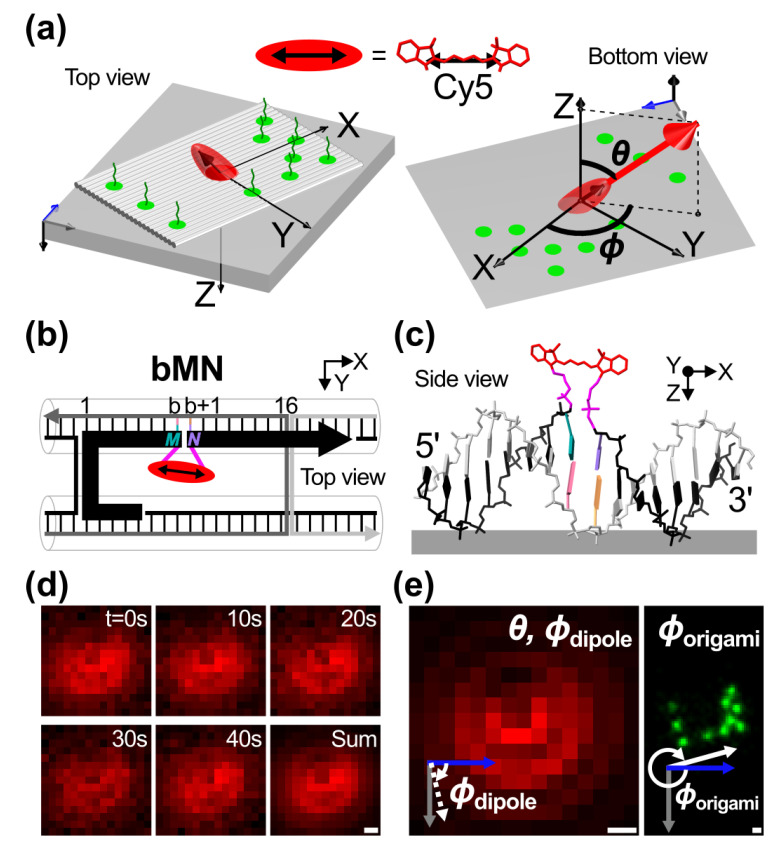
Figure 1.
Spatial orientation of a single Cy5 relative to the DNA origami template using dipolar imaging and DNA-PAINT. (a) Left: schematic of the template on the glass substrate. Red ellipsoid and green circles represent Cy5 and docking sites for DNA-PAINT imagers, respectively. The TDM of the dye is depicted as the double-headed arrow inside the ellipsoid. Right: Bottom view through the glass. The TDM orientation is given by the polar (θ) and azimuthal (ϕ) angles relative to the template. (b) Inset of the design of the template. A segment of the scaffold (thin black strand) is replaced by a Cy5-internally-labeled DNA (thick black strand) that is complementary to the dark grey staple. Cy5 is placed b number of bases from a common crossover. The position of the Cy5 was changed by tuning the sequences of the dark and light grey staples while keeping the same bases, including the M and N flanking bases. A general sample was named bMN. The replaced segment of the scaffold is not shown. (c) Schematics of the molecular structure of the dye tethered to the DNA duplex on glass. Both tethers are shown in pink. Neighboring bases are shown in different colors. (d) Representative frames of the dipole radiation pattern of 8TT (b = 8, M = T, N = T). Each frame was acquired for 2 s. Frames were summed to improve the signal-to-noise ratio. (e) Left: Simulated dipole radiation pattern with θdipole (=35°) and ϕdipole (=77°) at 550 nm defocused distance obtained after fitting the summed frame in (d). Right: DNA origami orientation, ϕorigami (= 345°), was obtained relative to the camera coordinates after DNA-PAINT reconstruction. The dye orientation was obtained as θ = θdipole and ϕ = ϕdipole-ϕorigami. The Cartesian coordinates of the origami and glass are the XYZ axes and the unlabeled blue, grey, and black arrows. Scale bars for the images of dipole radiation patterns and DNA-PAINT reconstruction are 200 nm and 10 nm, respectively.