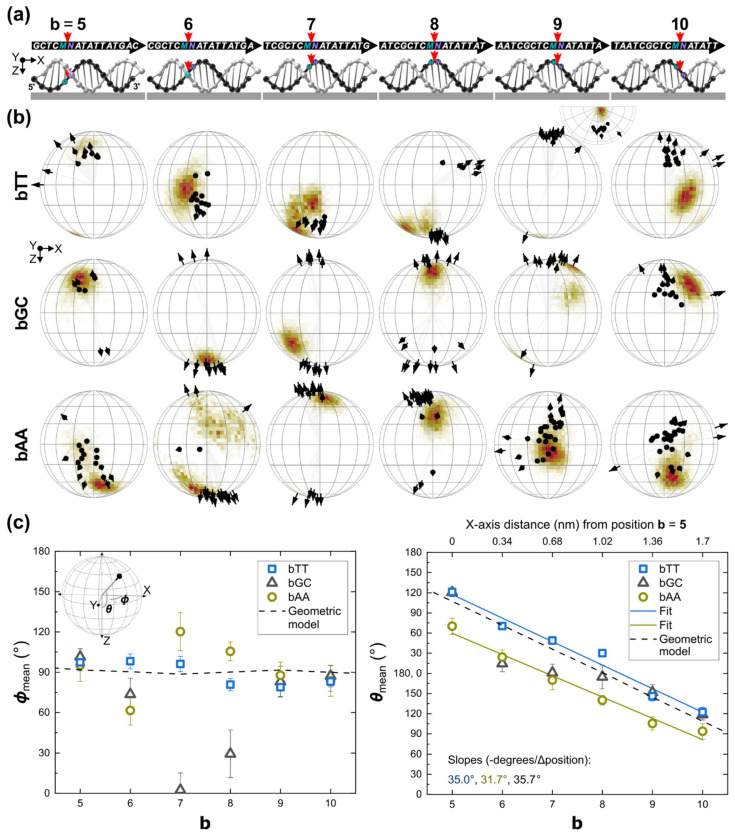
Figure 2.
The effect of the neighboring bases on the spatial orientation of Cy5, as a function of the attachment position b. (a) Schematics of DNA duplex on the glass substrate indicating b locations used in the study. DNA sequences are shown on top. Samples are labeled as bMN, where M and N were the dye’s flanking bases at the 5′ and 3′ ends, respectively. The 5′-to-3′ direction of the Cy5-tethered strand runs along the positive X-axis (see Figure 1a). (b) Results for bTT (top row), bGC (middle row) and bAA (bottom row). Each column of spheres corresponds to the schematics in (a). Experimental orientations are plotted on the unit hemisphere as black arrows. MD simulation results are plotted as histograms on the surface of the hemisphere. (c) Plots of ϕmean vs. b and θmean vs. b. The inset on the left shows again the coordinates for convenience. A geometric model (dash lines) was constructed based on the neighboring base pairs in a static DNA duplex (see Text S4.1). Error bars are , where and are the standard deviations of the elliptical cone with the center in the mean direction in the framework of the Kent (Fisher-Bingham) distribution [59].