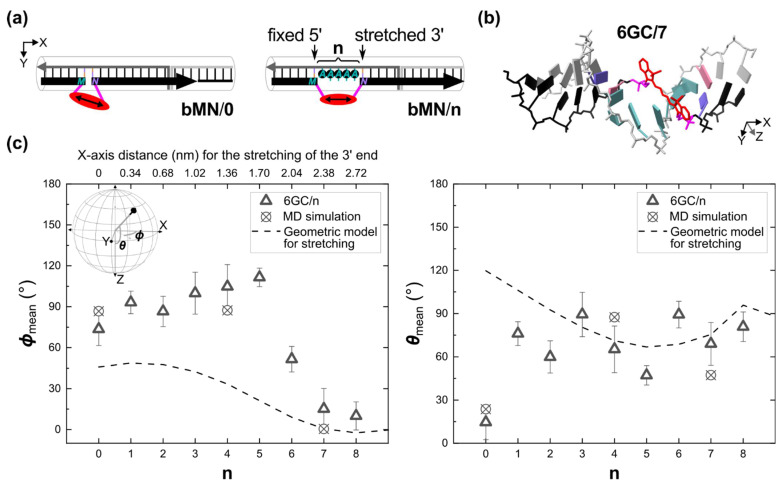
Figure 4.
The effect of mechanical stretching on the spatial orientation of Cy5, as a function of the stretching level n. (a) Schematics for the mechanical stretching using the same DNA template depicted in Figure 1b. Stretching was done in a stepwise manner by introducing n number of unpaired adenine bases. Samples are labeled as bMN/n, where M and N were the dye’s flanking bases at the 5′ and 3′ ends, respectively. The positive sign indicates that the 5′-to-3′ direction of the Cy5-tethered strand runs along the positive X-axis (see Figure 1a). Left: Structure without stretching, bMN/0. Right: Structure with stretching. (b) MD simulation frame of 6GC/7. (c) Plots of ϕmean vs. n and θmean vs. n for 6GC/n. The inset on the left shows again the coordinates for convenience. A geometric model (dash lines) was constructed based on the neighboring base pairs in a static DNA duplex (see Text S4.2). MD simulations for n = 0, 4, and 7 are shown as circles with x. Error bars are , where and are the standard deviations of the elliptical cone with the center in the mean direction in the framework of the Kent (Fisher-Bingham) distribution [59].