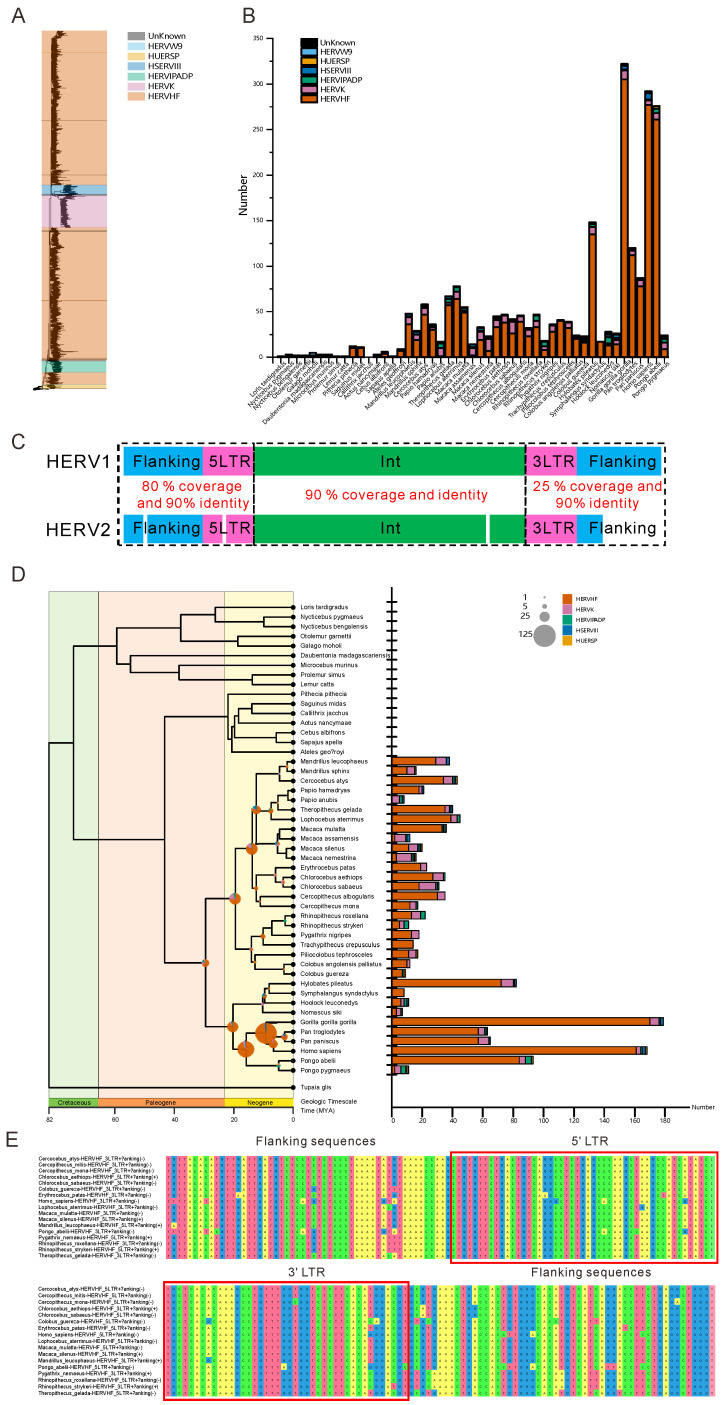
Figure 1.
The distribution, classification, and vertical transmission of F-HERVs in 49 primates. (A) The phylogenetic tree of all F-HERVs identified. The classifications of F-HERVs are indicated in different colors, and “Unknown” represents F-HERVs that cannot be classified according to their phylogenetic relationships with reference sequences or the similarity of their RT domains with reference RT sequences. Ultrafast bootstrap approximation (UFBoot) values over 95 are provided beside the nodes. (B) Histogram showing the number of F-HERVs in each species, and the classifications of HERVs are indicated in different colors. (C) The diagram shows the standards that were used to screen vertical transmission events. The blue boxes, pink boxes, and green boxes represent the flanking sequences (Flanking), LTR sequences (5LTR or 3LTR), and internal sequences (Int) of F-HERVs, respectively. The dashed boxes indicate the BLASTN searches we performed and the thresholds of each search (in red). The white segments represent the mismatch or gap in BLASTN. The detailed procedure of vertical transmission identification is provided in the Materials and Methods. (D) The left panel shows the rooted phylogenetic tree and the divergence times of 50 primates (Tupaia glis was used as the root), and the numbers of possible vertical transmission events are indicated on the corresponding nodes by the area of pies. The right panel shows the numbers of vertical transmission-related F-HERVs in each species we studied. The classifications of the F-HERVs are indicated in different colors. (E) The image shows the alignments of the 5′LTR and flanking sequences (upper) and the 3′LTR and flanking sequences (lower) of a widely vertically transmitted F-HERV-H in 17 species. The names of the species are listed on the left, and red boxes indicate the LTR sequences of F-HERVH. The full alignments of the LTR and flanking sequences are provided in the Supplementary Materials.