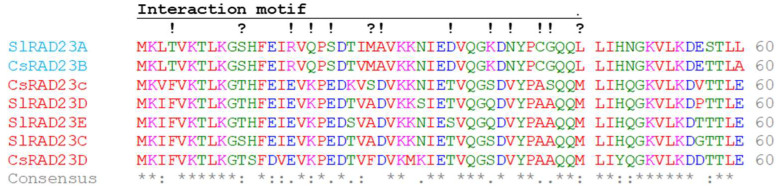
Figure 4.
Alignment of the S. lycopersicum and C. sinensis RAD23 proteins. The names of the sequences in Clade 1 are in blue; those in Clade 2 are in red. Amino acid residues are colored according to their physiochemical properties: red = small (small + hydrophobic including aromatic − Y), blue = acidic, magenta = basic − H, and green = hydroxyl + sulfhydryl + amine + G. Consensus symbols are as follows: an asterisk (*) indicates the positions which have a single, fully conserved residue; a colon (:) indicates conservation between groups of strongly similar properties; and a period (.) indicates conservation between groups of weakly similar properties. The motif interacting with Lso-HPE1 is identified using a line; the symbols below the line are as follows: a question mark (?) indicates the positions with amino acid substitutions with similar physiochemical properties between clades, and an exclamation mark (!) indicate the positions with amino acid substitutions with different physiochemical properties between clades.