FIGURE 1.
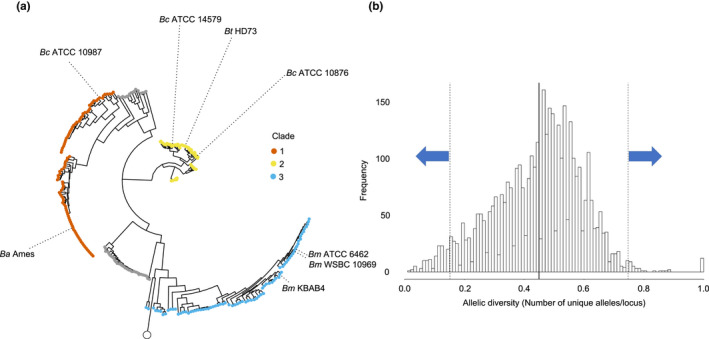
Identifying core and accessory genes under selection within the Bacillus cereus sensu lato (Bc sl) phylogeny. (a) Maximum‐likelihood phylogeny of the Bc sl group strains used in this study. The concatenated core genome sequences were aligned using mafft and fed into iq‐tree. Clade identity was determined through reference to type strains and consultation of existing clade metadata. (b) The process by which genes of low and high diversity were identified. The graph shows the frequency distribution of allelic diversity values across genes within a clade's flexible core genome. The solid line shows the mean diversity of the strict core genome within the clade and the dashed lines show the second standard deviation interval of the strict core genome
