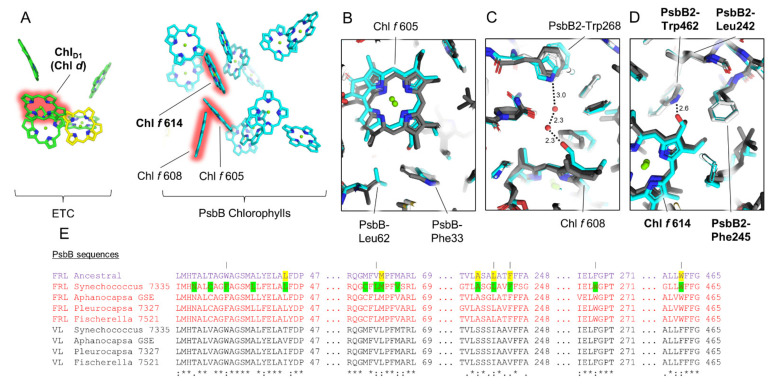
Figure 6.
Conservation of FRL-PsbB interactions near the Chl f molecules bound to it. In (A), the Chls and pheophytins in the electron transfer chain and FRL-PsbB of the Synechococcus 7335 apo-FRL-PSII structure are shown as tetrapyrrole rings only from a membrane plane view. The Chls d and f are highlighted in red. In (B–D), the Synechococcus 7335 apo-FRL-PSII structure (cyan), the homology model of the FRL-PsbB ancestral sequence (white), and two non-FaRLiP holocomplex PSII structures (light and dark grey from T. vulcanus and Synechocystis 6803, respectively) are superimposed. (B–D) show the regions of the structures near the C2 formyl moiety of the three Chl f molecules bound to FRL-PsbB (sites 605, 608, and 614 based on the original assignment of Chl a sites from T. vulcanus PSII [46]), respectively. They also show H-bonding interactions involving the C2 formyl moieties of Chl f molecules in dashed lines with distances in units of Å where applicable. In (A–D), features that appear to be conserved in the ancestral FRL-PSII are labeled in bold font. In (E), a series of partial sequence alignments is shown that includes the FRL ancestral sequence and FRL- and VL-specific sequences from extant FaRLiP-capable cyanobacteria. Conserved FRL-specific residues in extant cyanobacteria are highlighted in green in the sequence from Synechococcus 7335. If the same position is conserved in the FRL ancestral sequence, it is highlighted in yellow. Vertical lines above residue positions in (E) correspond to amino acids labeled in (B–D). The Clustal Omega sequence conservation identifiers are shown below the alignment.