Abstract
Hepatocellular carcinoma (HCC) is the most common type of primary liver cancer. Metabolic reprogramming is considered to be an important hallmark of cancer. Emerging studies have demonstrated that noncoding RNAs (ncRNAs) are closely associated with metabolic reprogramming of HCC. NcRNAs can directly regulate the expressions or functions of metabolic enzymes or indirectly regulate the metabolism of HCC cells through some vital signaling pathways. Until now, the mechanisms of HCC development and progression remain largely unclear, and understanding the regulatory mechanism of ncRNAs on metabolic reprogramming of HCC may provide an important basis for breakthrough progress in the treatment of HCC. In this review, we summarize the ncRNAs involved in regulating metabolic reprogramming of HCC. Specifically, the regulatory roles of ncRNAs in glucose, lipid and amino acid metabolism are elaborated. In addition, we discuss the molecular mechanism of ncRNAs in regulation of metabolic reprogramming and possible therapeutic strategies that target the metabolism of cancer cells by modulating the expressions of specific ncRNAs.
Keywords: circRNA, hepatocellular carcinoma, lncRNA, metabolic reprogramming, noncoding RNA
Abbreviations
- 1,3‐BPG
1,3‐bisphosphoglyceric acid
- α‐KG
α‐ketoglutarate
- ABCA1
ATP‐binding cassette transporter A1
- ACC
acetyl‐CoA carboxylase
- ACLY
ATP‐citrate lyase
- ACSL1
acyl CoA synthetase long‐chain 1
- AGO2
argonaute 2
- ALDOA
aldolase
- AMPK
adenosine 5‐monophosphate‐activated protein kinase
- ASCT2
alanine‐serine‐cysteine transporter 2
- ATGL
adipose triglyceride lipase
- BCAA
branched‐chain amino acid
- BCAT1
branched‐chain amino acid transaminase 1
- BCKA
branched‐chain keto acid
- BCKD
branched‐chain ketoacid dehydrogenase
- CAT‐1
cationic amino acid transporter 1
- CCT3
chaperonin containing TCP1 subunit 3
- CS
citrate synthase
- DHAP
dihydroxyacetone phosphate
- ENO1
enolase 1
- ERK1
extracellular regulated protein kinases 1
- ERK2
extracellular regulated protein kinases 2
- F‐1,6‐P
fructose‐1,6‐bisphosphate
- F‐6‐P
fructose‐6‐phosphate
- FASN
fatty acid synthase
- G‐3‐P
glyceraldehyde 3‐phosphate
- G‐6‐P
glucose‐6‐phosphate
- G6PC
glucose‐6‐phosphatase
- G6PD
glucose‐6‐phosphate dehydrogenase
- GAPDH
glyceraldehyde phosphate dehydrogenase
- GLS1
glutaminase 1
- GLUT
glucose transporter
- HCC
hepatocellular carcinoma
- HDL
high‐density lipoprotein
- HK2
hexokinase 2
- HMGCR
3‐hydroxy‐3‐methylglutaryl CoA reductase
- HULC
highly upregulated in liver cancer
- IDH
isocitrate dehydrogenases
- IGF‐1R
insulin‐like growth factor receptor‐1
- LDHA
lactate dehydrogenase A
- LXR
liver X receptor
- MAGL
monoacylglycerol lipase
- MALAT1
metastasis‐associated lung adenocarcinoma transcript 1
- MAT
methionine adenosinetransferase
- MDH
malate dehydrogenase
- PC
pyruvate carboxylase
- PCK1
phosphoenolpyruvate carboxykinase 1
- PDH
pyruvate dehydrogenase
- PDK1
pyruvate dehydrogenase kinase 1
- PEP
phosphoenolpyruvate
- PFK
phosphofructokinase
- PGK1
phosphoglycerate kinase 1
- PI3K
phosphoinositide 3‐kinase
- PKM1/M2
pyruvate kinase isozymes M1/M2
- PPP
pentose phosphate pathway
- PTEN
phosphatase and tensin homolog
- SAM
sadenosyl‐l‐methionine
- SCD
stearoyl‐CoA desaturase
- SCS
succinyl coenzyme A synthetase
- SDH
succinate dehydrogenase
- SIRT1
sirtuin 1
- SREBF
sterol regulatory element‐binding protein
- SREBPs
sterol regulatory element binding proteins
- SRSF1
serine/arginine‐rich splicing factor 1
- TCA
tricarboxylic acid cycle
1. INTRODUCTION
Liver cancer is now the second leading cause of cancer death worldwide. 1 HCC is the main form of primary liver cancer. 2 The metabolic pattern of the cancer cells changes significantly during tumorigenesis, which is known as metabolic reprogramming. Increasing evidence show that metabolic reprogramming plays an essential role in HCC occurrence and metastasis. To some extent, HCC is a kind of metabolic disease. Cancer cells metabolize glucose primarily through glycolysis, whether they have enough oxygen or not, 3 , 4 which have sparked a boom in cancer metabolism research for decades. 5 HCC cells can quickly transform their glycometabolism from the oxidative phosphorylation metabolic pathway to the glycolysis pathway in hypoxia conditions, known as the Warburg effect. This effect not only provides a favorable microenvironment for tumor progression, but also meets the need for rapid cell proliferation. HCC cells also require lipid energy for growth and membrane synthesis, and modification in lipid metabolism enables cancer cells to survive hypoxia, and modifications in lipid metabolism enable HCC cells to survive hypoxia and drug treatment. In addition, with the rapid growth of hepatoma cells, the demand for amino acids also increases to support the rapid proliferation of cancer cells and maintain oxidative homeostasis. In brief, reprogramming of HCC cells by changing the mode of material metabolism is a metabolic marker that supports hepatocarcinogenesis.
Over the past many years, ncRNAs, which refer to RNAs that do not encode proteins, have been considered as “noise” in the processes of transcription. 6 The importance of ncRNAs in biological events has been recognized now. Less than 2% of the genes encode proteins in human genome, the vast majority of the rest genes are transcribed into ncRNAs. Thus, some researchers predict that ncRNAs play a significant role as proteins in the biological processes. In the past decade, ncRNAs have attracted extensive attention due to their novel and important biological regulatory capabilities. There is growing evidence 7 , 8 , 9 , 10 that ncRNAs are altered during the development and progression of HCC and play crucial regulatory roles as oncogenes or tumor suppressor genes (Table S1). For instance, miR‐155 is upregulated in HCC and can promote the progression of HCC by activating Wnt signaling; miR‐106b can boost cell proliferation and tumor growth of HCC. 11 , 12 Besides, miR‐125b and miR‐504 promote hepatocarcinogenesis by regulating the expression of p53, 13 , 14 miR‐125a suppresses the progression of HCC through inhibiting the PI3K/AKT/mTOR signaling pathway. 15 Furthermore, the deregulation of long noncoding RNAs (lncRNAs) has been detected in HCC 16 (Table S1). Recent studies have found that the expression of linc01146 in HCC was decreased and associated with the prognosis of HCC patients. Additionally, bioinformatic analysis revealed that linc01146 might be associated with metabolic pathways. 17 It was found that lncCYTOR promoted the proliferation of HCC by regulating the miR‐125a‐5p/LASP1 axis, 18 lncH19 was upregulated and carcinogenic under hypoxia, 19 lncRNA ATB has been shown to promote EMT and metastasis of HCC by upregulating TGF‐β. 20 In addition to lncRNAs, dysregulation of circRNAs in HCC has been found in numerous studies in recent years (Table S1). For instance, circRHOT1 has been found to significantly promote HCC growth and metastasis by initiating NR2F6 expression, 21 and circSMARCA5 have been demonstrated to inhibit the growth and metastasis of HCC by sponging miR‐17 and miR‐181b. 22 A recent study identified that circMRPS35 promoted HCC progression by regulating the expression of Syntaxin 3 via sponged microRNA‐148a‐3p, 23 another recent study found that circKCNN2 inhibited HCC recurrence through the miR‐520c‐3p/MBD2 axis and might be a promising biomarker for predicting HCC recurrence. 24
HCC cells are often modulated by genetic changes and tumor microenvironmental stress. 25 During the development and progression of HCC, the metabolic pathways of cancer cells are often regulated by ncRNAs. Metabolomics studies provide new insights into the mechanisms of metabolic reprogramming and offer a promising individualized therapeutic strategy in HCC. 26 , 27 Therefore, it is of great significance to study the role and mechanism of ncRNAs in HCC cell metabolism. In this review, we introduce the classification of ncRNAs, summarize the dysregulated ncRNAs in HCC and describe the ncRNAs involved in metabolic reprogramming in HCC. Moreover, we discuss the molecular mechanism and role of ncRNAs in regulating the metabolism of glucose, lipid and amino acids in HCC.
1.1. Classification of ncRNAs
A variety of RNAs have been found in human beings. Generally, they can be divided into coding RNAs and ncRNAs according to their protein‐coding capacity. 28 , 29 The coding RNAs include mRNAs, while the ncRNAs include many types, such as microRNA (miRNA), long noncoding RNA (lncRNA), small nuclear RNA (snRNA), small nucleolar RNA (snoRNA), piwi‐interacting RNA (piRNA), circular RNA (circRNA), small interfering RNA (siRNA), signal recognition particle RNA (srpRNA) and so on. 30 , 31 , 32 , 33 , 34 According to their size, ncRNAs can be divided into lncRNAs and small noncoding RNAs (sncRNAs). lncRNAs refer to ncRNAs with a length greater than 200 nucleotides, while sncRNAs are ncRNAs that are less than 200 nucleotides, 35 , 36 such as miRNAs, siRNAs, piRNAs, circRNAs, snRNAs, snoRNAs, srpRNAs and telomerase RNAs. 37 , 38 , 39
1.2. ncRNAs and metabolic reprogramming in HCC
In recent years, ncRNAs have been proved to play a critical role in various cellular activities, including material metabolism, gene activation and silencing, RNA splicing, modification and editing and protein transport and translation. 40 , 41 , 42 Most notably, they are widely known as key regulators of cancer metabolism. 43 , 44 , 45 There is increasing evidence that ncRNAs are involved in regulation of metabolic reprogramming in HCC. Among them, miRNA is the most studied in regulation of metabolic reprogramming in HCC. A large number of miRNAs were found to be closely related to HCC metabolic reprogramming, such as miR‐122, miR‐195, miR‐451, miR‐33, miR‐34a and miR‐520 (Table S2). Additionally, lncRNAs have also been found to be extensively involved in the metabolic reprogramming of HCC in recent years, such as lncHULC, linc01554, lncRNA‐p21, lncMVIH, lncMALAT1, lncWFDC21P, lncRP11‐241J12.3 and linc00326 (Table S3).
Aside from miRNAs and lncRNAs, the functions of circRNAs are also gaining increasing attention. CircRNAs are the latest research hotspot in ncRNA field in recent years. Sanger et al 46 proposed the term “circRNA” for the first time more than 40 years ago, and showed that circRNA has a closed ring structure and more stable expression, and is hard to degrade and is not affected by RNA exonucleases. The circRNAs are produced by “back splicing” of mRNAs or linear ncRNAs. 47 Many circRNAs play essential biological functions by acting as miRNAs “sponges” to regulate protein function or selftranslation. 48 , 49 , 50 Up to now, more than 100 000 human circRNAs have been discovered, 51 , 52 but their functional roles are mostly unknown. Most studies of circRNAs have focused on their roles in tumors, and only a few biological functions of circRNAs have been studied. 49 , 50 , 53 , 54 It was found that circCDR1as, circSMARCA5, circ‐ZEB1.33, circSETD3, circMTO1, circSMAD2, circFBLIM1 and circHIPK3 played oncogenic or suppressive roles in the progression of HCC. 22 , 55 , 56 , 57 , 58 , 59 , 60 , 61 Furthermore, a series of circRNAs were found to be closely related to metabolic reprogramming in HCC, 62 such as circMAT2B, circSPECC1, circC3P1 and circRPN2 63 , 64 , 65 , 66 (Table S2).
To date, there are relatively few studies on the role of small nucleolar RNAs (snoRNAs) and Piwi‐interacting RNAs (piRNAs) in the metabolic reprogramming of HCC. SnoRNAs are a class of noncoding small RNAs ranging in length from 60 to 300 nt, and can bind with nucleolar ribonucleoproteins to form snoRNPs complexes. 67 The biological processes in which snoRNAs are involved mainly include rRNA processing, regulation of RNA splicing and translation and oxidative stress response. 68 The structure of snoRNAs is very conservative and can be divided into two categories: C/D box snoRNAs and H/ACA box. 69 , 70 , 71 , 72 Recently, we have found that two snoRNAs are involved in the metabolism of HCC, namely snoRD113‐1 and snoRD126 (Table S2). PiRNAs are a kind of small RNA molecules, with a length of about 24 to 31 nucleotides and a rich variety of up to 15 000 species. 73 They can bind with PIWI protein to form piRNA complexes (piRCs) and are related to RNA silencing. PiRNAs mainly act on mammalian germ cells and regulate transposon expression. 74 , 75 A study published 5 years ago identified the expression characteristics of 125 piRNAs associated with HCC, 39 but studies on the roles of piRNA in HCC metabolism are rare.
Collectively, ncRNAs are extensively involved in metabolic reprogramming of HCC (Table S2).
2. ncRNAs AND GLUCOSE METABOLISM IN HCC
2.1. miRNAs and glucose metabolism
Among ncRNAs, miRNAs are currently the most well studied. 76 They are a class of noncoding single stranded RNA molecules, which are involved in the regulation of post‐transcriptional gene expression. There are two main ways in which miRNA participates in gene regulation: promoting mRNA degradation or inhibiting protein translation. 77 , 78 In the recent decade, miRNA expression profiles in HCC have been widely studied. Glucose metabolism mainly includes glycolysis, tricarboxylic acid cycle and pentose phosphate pathway. Glycolysis is often upregulated in HCC, and a remarkable feature of HCC cells is enhanced glucose uptake, 79 which greatly accelerates the decomposition of glucose whether in aerobic or hypoxic environment. 80 Studies have demonstrated that metabolic reprogramming predates HCC in mouse hepatocytes. 81 A growing number of evidence suggests that changes in tumor suppressor genes and oncogenes, via regulating the effectors of key metabolic enzymes, are responsible for metabolic reprogramming. 82 Next, the role of miRNA in glucose metabolism will be discussed in detail.
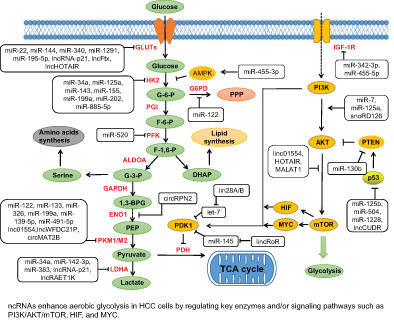
As a matter of fact, numerous studies have found that miRNA promotes glucose metabolism of HCC by regulating a variety of glycolytic factors or key enzymes. 83 Pyruvate kinase (PK), which is one of the most widely studied enzymes in glycometabolism, is divided into M‐type and L‐type, and there are two types of M‐type, M1 and M2. In HCC cells, it has been reported that increasing miRNAs modulate the expression of PKM (a rate‐limiting enzyme of glycolysis). miR‐199a is inhibited under hypoxia and hinders glycolysis of HCC cells by targeting pyruvate kinase‐M2 (PKM2). 84 Another study has shown that cholesterol modified agomiR‐199a can restrain the uptake of [(18)f]‐fluorodeoxyglucose and growth of HCC in mice. 85 Moreover, miR‐122, which is the most aplenty miRNA in the liver, 86 restrains glycolysis of HCC cells by targeting PKM2. 87 , 88 Meanwhile, it also reduces the incidence of HCC metastasis by downregulating PKM2. 89 In HCC cell lines, miR‐326 also inhibits PKM2 and has potential antitumor effects. 90 HK, another key enzyme in glucose metabolism, is also extensively regulated by miRNAs. There are four isozymes of HK in human body. Of them, HK1 is mainly distributed in brain; HK2 is mainly distributed in skeletal muscle; HK3 is mainly distributed in leukocytes; and HK4 is mainly distributed in liver. HK4 is also called glucokinase owing to its strong specificity to glucose, which is quite different from the other three isoenzymes. A study found that hypoxia induces downregulation of miR‐125a expression and directly targets HK2, thereby inhibiting HCC glycolysis (eg, reducing glucose uptake and production of lactic acid and ATP in HCC cells). 91 Moreover, overexpression of miR‐202 has also been reported to inhibit HCC proliferation and glucose metabolism notably (such as the uptake of glucose and lactic acid production) by directly targeting HK2. 92 Another study indicated that upregulation of miR‐885‐5p could significantly reduce glucose uptake and lactate production of HCC cells by targeting 3′UTR of HK2 directly. In the meantime, it suppressed the migration and proliferation of HCC cells, and promoted cell apoptosis in vitro and tumor growth in vivo. 93 What's more, miR‐33b regulates glucose metabolism by targeting glucose‐6‐phosphatase (G6PC, a key enzyme of hepatic gluconeogenesis) and phosphoenolpyruvate carboxykinase 1 (PCK1). In human hepatocytes, overexpression of miR‐33b suppresses the expression of G6PC and PCK1 and results in a significant decrease in glucose production. 94 A recent study identified a mitochondrial miRNA (mitomiR‐181a‐5p) that targets the genes encoding mt‐CYB and mt‐CO2. Overexpression of mitomiR‐181a‐5p decreased mt‐CYB and mt‐CO2 levels in HCC cells and upregulated the expression of HK2 and GLUT1. Further studies have found that overexpression of mitomiR‐181a‐5p causes glucose metabolic reprogramming and promotes HCC growth and metastasis. 25 miR‐34 and let‐7 regulate glycolysis of HCC cells through targeting lactate dehydrogenase A (LDHA) via p53. 95 MiR‐375 is related to the function of islet cells, and the deletion or overexpression of miR‐375 will affect glucose metabolism. 96 All in all, dysregulation of miRNA could lead to HCC metabolic reprogramming by targeting the expression of metabolic enzymes directly (Figure 1).
FIGURE 1.
ncRNAs regulate glucose metabolism in HCC. Regulation of glucose metabolism by ncRNAs in HCC cells (Tables S2 and S3). ncRNAs enhance aerobic glycolysis in HCC cells by regulating key enzymes and/or signaling pathways such as PI3K/AKT/mTOR, HIF and MYC. The major metabolites are shown in green, the main metabolic enzymes are shown in red and the main metabolic pathways are indicated in yellow. 1,3‐BPG, 1,3‐bisphosphoglyceric acid; ALDOA, aldolase; DHAP, dihydroxyacetone phosphate; F‐1,6‐P, fructose‐1,6‐bisphosphate; F‐6‐P, fructose‐6‐phosphate; G‐3‐P, glyceraldehyde 3‐phosphate; G‐6‐P, glucose‐6‐phosphate; G6PD, glucose‐6‐phosphate dehydrogenase; GAPDH, glyceraldehyde phosphate dehydrogenase; GLUT, glucose transporter; HK2, hexokinase 2; IGF‐1R, insulin‐like growth factor receptor‐1; LDHA, lactate dehydrogenase A; PDH, pyruvate dehydrogenase; PDK1, pyruvate dehydrogenase kinase 1; PEP, phosphoenolpyruvate; PFK, phosphofructokinase; PKM1/M2, pyruvate kinase isozymes M1/M2; PPP, pentose phosphate pathway [Color figure can be viewed at wileyonlinelibrary.com]
Many studies have found that lots of signaling pathways involved in glucose metabolism are indirectly regulated by miRNAs. The main indirect pathways involved in metabolic reprogramming include AMPK, AKT, MYC and HIF signaling pathways. Adenosine 5′‐monophosphate (AMP)‐activated protein kinase (AMPK), a crucial kinase regulating energy homeostasis, is a key protein involved in a variety of signaling pathways. One study indicated that miR‐455‐3p directly targets AMPK and regulates HCC cell growth, metastasis and glycolysis by modulating HK2 activity. 97 AKT, also known as PKB or Rac (there are three isoforms: AKT1, AKT2 and AKT3), is a serine/threonine protein kinase. AMPK can activate AKT by regulating phosphatidylinositol‐3 kinase (PI3K), 98 which is a family of proteins that play a principal role in metabolism. The PI3K/AKT pathway is regulated by several miRNAs. 99 For instance, miR‐21, miR‐337 and miR‐130b induce angiogenesis of HCC by targeting phosphatase and tensin homolog (PTEN, a key phosphatase) and activating the PI3K/AKT signaling pathway. 100 , 101 , 102 MiR‐7 inhibits the progression of HCC by downregulating the PI3K/AKT/mTOR and IGF‐IR/AKT signaling pathways. A further investigation shows overexpression of miR‐7 can inhibit glucose metabolism, tumor invasion and metastasis in HCC. 103 , 104 mTOR is a key kinase downstream of PI3K/AKT, which regulates tumor cell proliferation, growth, survival and angiogenesis. Another study has indicated that MYC promotes the conversion of pyruvate to alanine in the early stages of HCC and to lactic acid in the late stages. 105 MiR‐342‐3p and miR‐455‐5p inhibit HCC proliferation and invasion by suppressing IGF‐1R‐mediated Warburg effect. 106 , 107 Similarly, miR‐3662 suppresses HCC growth via inhibition of HIF‐1α‐mediated reprogramming of glucose metabolism. 108 In summary, miRNAs are directly or indirectly involved in the metabolic reprogramming of HCC cells (Figure 1).
2.2. Other ncRNAs and glucose metabolism
Recent studies have found that in addition to a burst of miRNAs involved in glucose metabolism of HCC, many other ncRNAs have also been found to be involved in the regulation of glucose metabolism in HCC, such as lncRNAs, circRNA, piRNA, snoRNAs and so on (Figure 1).
Okazaki et al identified a new class of ncRNAs 20 years ago, later known as lncRNAs. 109 LncRNAs interact with various protein factors, so they can play a regulatory role at transcriptional level or post‐transcriptional level. 45 , 110 , 111 , 112 In HCC, lncRNAs mainly regulate gene expression and metabolic changes via interacting with RNA and proteins. 39 Recent studies have found that quite a few lncRNAs are also involved in regulation of metabolic reprogramming in HCC. A study found that linc01554 inhibited hepatocarcinogenesis by downregulating PKM2 expression and suppressing AKT/mTOR pathway to restrain aerobic glycolysis in HCC. 113 Interestingly, knockout of linc01554 could effectively reverse the tumor inhibition of linc01554. Yang et al 114 revealed that lncRNA‐p21 was a hypoxic reactive lncRNA, which was critical for hypoxia enhanced glycolysis. lncRNA‐p21 could modulate HIF‐1α transcriptional activity and promote glucose uptake and lactic acid production by regulating HIF‐1α expression level under hypoxia. Knockout of lncRNA‐p21 reduced the activity of LDHA and the expression level of LDHA and GLUT1, suggesting that lncRNA‐p21 promoted glycolysis under hypoxia. 114 Moreover, a recently published study indicated that lncRP11‐241J12.3 promoted HCC growth and aggressiveness by upregulating the expression of pyruvate carboxylase (PC) and MSH3 (a key protein in pyruvate metabolism) 115 (Figure 2). Through genome‐wide screening analysis of HCC cohorts in the TCGA database, Wang et al 116 found that five metabolism‐related lncRNAs (AC099850.3, AL031985.3, AL365203.2, LUCAT1 and MIR210HG) had independent prognostic significance in predicting the clinical prognosis of HCC patients and were associated with overall survival of patients. These lncRNAs can be used as potential biomarkers of HCC, but how to regulate the metabolism of HCC remains to be further studied. Another study has confirmed that lncRNA MVIH promotes HCC growth and intrahepatic metastasis by inhibiting the secretion of phosphoglycerate kinase 1 (PGK1, an important glycolytic enzyme). Further research revealed MVIH expression was negatively correlated with serum PGK1 level in HCC patients. 117 , 118 Additionally, it is reported that lncMALAT1 enhances the translation of metabolic transcription factor TCF7L2 by upregulating serine/arginine‐rich splicing factor 1 (SRSF1) and activating the mTORC1‐4EBP1 axis, thus upregulating the expression of glycolytic genes and downregulating gluconeogenic enzymes, indicating that MALAT1 promotes the development and progression of HCC by reprogramming glucose metabolism. 119
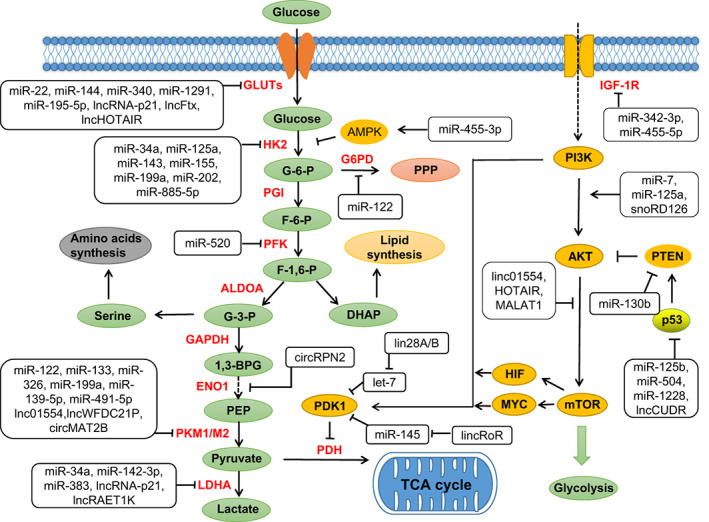
FIGURE 2.
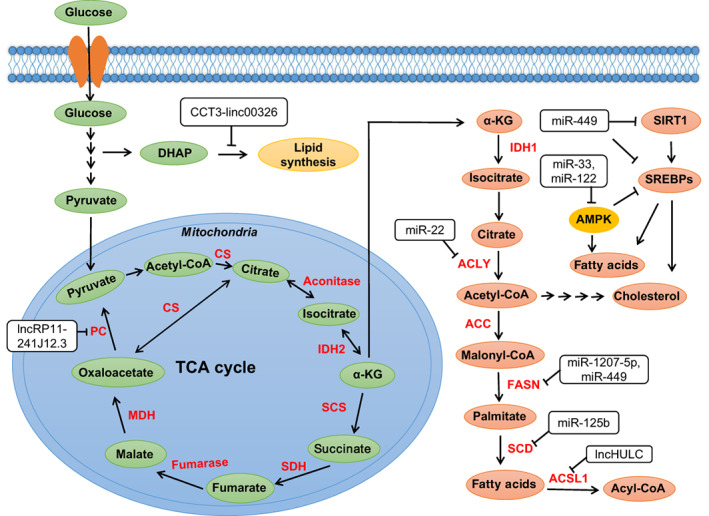
ncRNAs regulate the steps of TCA cycle and lipogenesis in HCC. ncRNAs regulate TCA cycle and lipogenesis by regulating key enzymes of lipid metabolism and some crucial proteins such as AMPK, SIRT1 and SREBPs (Tables S2 and S3). α‐KG, α‐ketoglutarate; ACC, acetyl‐CoA carboxylase; ACLY, ATP‐citrate lyase; ACSL1, long‐chain acyl CoA synthetase 1; CS, citrate synthase; FASN, fatty acid synthase; IDH, isocitrate dehydrogenases; MDH, malate dehydrogenase; SCD, stearoyl‐CoA desaturase; SCS, succinyl coenzyme A synthetase; SDH, succinate dehydrogenase; IRT1, sirtuin 1; SREBPs, sterol regulatory element binding proteins [Color figure can be viewed at wileyonlinelibrary.com]
Notably, recent studies have found that some circRNAs are also involved in the regulation of glucose metabolism in HCC. The circMAT2B upregulates PKM2 expression and enhances glycolysis through sponging miR‐338‐3p under hypoxia, thus promoting the progression of HCC. 63 Under oxidative stress, circSPECC1 promotes hepatocarcinogenesis by directly suppressing the expression of TGF‐β2, which is proved to be the target gene of miR‐33a. Furthermore, circSPECC1 was significantly downregulated under H2O2 treatment, and knockdown of it inhibited HCC cell proliferation and promoted apoptosis. 64 A recently published study demonstrated that circRPN2 inhibited HCC aerobic glycolysis and metastasis via accelerating enolase 1 (ENO1) degradation and modulating the miR‐183‐5p/FOXO1 axis. 66
Not only that, Law et al 120 found that a new piRNA (piR‐Hep1) was upregulated in HCC in a small RNA transcriptome sequencing analysis. Further studies indicated that silencing piR‐Hep1 could decrease AKT phosphorylation and inhibit the growth and aggressiveness of HCC. Furthermore, research has found that snoRD113‐1 is downregulated in HCC and can reduce the survival rate of patients. 39 Another study confirmed that snoRD113‐1 inhibited HCC growth by restraining phosphorylation of extracellular signal‐regulated kinases (ERK) 1 and ERK2. 121 In addition, snoRD126 is upregulated in HCC and promotes the growth of HCC by upregulating genes in the PI3K‐AKT pathway. 122
2.3. ncRNAs and lipid metabolism
Compared to glucose metabolism, lipid metabolism has rarely been reported in the past few years, but with extensive research in the fatty acid metabolism, the role of lipid metabolic reprogramming in cancer cells is becoming increasingly apparent. 123 Lipids mainly include triglycerides and lipoids. Lipoids consist of phospholipids, glycolipids and cholesterol (esters). 124 The main functions of lipids in cell metabolism are energy storage, fatty acid synthesis, biomembrane formation and signal transduction. 125 Lipid metabolism of cancer cells is mainly reflected in the synthesis of lipids, storage of lipids and the use of cholesterol esters to promote the migration of tumor cells. The synthesis of fatty acids starts from acetyl‐CoA and NADPH, which are synthesized from the tricarboxylic acid (TCA) cycle and the pentose phosphate pathway (PPP) (Figure 2).
The lipid metabolism of HCC cells is regulated by some ncRNAs. MiR‐122 is an important regulator of liver fatty acid and cholesterol metabolism. Knockout or overexpression of miRNA‐122 in mice reduces or increases cholesterol biosynthesis, respectively. 96 Moreover, inhibition of miR‐122 increased fatty acid oxidation in mice liver, and also indirectly led to a decrease of cholesterol synthetase, which resulted in the decrease of cholesterol synthesis. 126 , 127 Another study has indicated that knockout of miRNA‐122 reduces blood cholesterol, liver cholesterol and fatty acid synthesis in mice, while increases liver fatty acid oxidation. 128 The study also found increased phosphorylation of AMPK in the liver of mice and suggested that miR‐122 might be involved in regulating AMPK signaling (Figure 2). The main direct targets of ncRNA during regulation of metabolic reprogramming in HCC include various lipases such as isocitrate dehydrogenases (IDH), ATP‐citrate lyase (ACLY), acetyl CoA carboxylase (ACC), fatty acid synthase (FASN), Stearoyl‐CoA desaturase (SCD), fatty triacylglycerol lipase (ATGL), monoacylglycerol lipase (MAGL) and 3‐hydroxy‐3‐methylglutaryl CoA reductase (HMGCR). MiR‐33 interferes with fatty acid degradation by binding to sterol regulatory element‐binding protein (SREBF) transcription factor, which inhibits the translation of fatty acid beta‐oxidase 129 (Figure 2). SREBPs are crucial transcription factors that regulate fatty acid and cholesterol metabolism. 130 , 131 There are three isoforms of SREBPs, namely SREBP‐1a, SREBP‐1c and SREBP‐2, which have different roles in adipogenesis. 132 miR‐449 was found to control adipogenesis and cholesterogenesis and inhibit the proliferation of HCC cells by suppressing the expression of SIRT1 (a NAD+‐dependent deacetylase that regulates a series of genes involved in lipids regulation) and SREBP‐1c and downregulating FASN and HMGCR (Figure 2). Furthermore, miR‐33a and miR‐33b regulate lipid metabolism in coordination with the SREBP 133 , 134 (Figure 2). Meanwhile, they were also found to regulate ATP‐binding cassette transporter A1 (ABCA1) to promote the conversion of intracellular free cholesterol to apolipoprotein A1 and high‐density lipoprotein (HDL) formation. 135 AMPK can suppress the expression of liver X receptor (LXR) and SREBP‐1. 136 Hence, miR‐33 downregulation of AMPK might alleviate AMPK suppression of SREBPs, thus enhancing lipid levels 137 , 138 (Figure 2).
In addition to miRNAs, other ncRNAs are also involved in lipid metabolism of HCC. Highly upregulated in liver cancer (HULC) is the first lncRNA found to be specifically upregulated in HCC. 139 , 140 It can promote the accumulation of triglycerides and cholesterol in HCC cells via miR‐9/PPARα/ACSL1 signaling pathway. The mechanism of our study suggested that HULC regulated abnormal regulation of lipid metabolism in HCC by activating acyl‐CoA synthase subunit ACSL1 and overexpression of ACSL1 promoted the proliferation of HCC cells (Figure 2). On the one hand, HULC activated the ACSL1 promoter in HCC cells by upregulating the transcription factor PPARA; on the other hand, HULC induced methylation of CpG islands in the miR‐9 promoter to inhibit miR‐9 targeting of PPARA mRNA. 141 Additionally, a recent study identified that perturbation of the CCT3‐LINC00326 axis reduced lipid accumulation and increased lipid degradation in HCC cells, as well as diminished tumor growth in vivo 142 (Figure 2).
2.4. ncRNAs and amino acid metabolism
The metabolism of amino acids in cells includes two aspects, anabolism and catabolism. The former refers to the synthesis of proteins, peptides and other nitrogen‐containing materials required by the body. The latter refers to the decomposition of α‐ketoacids, amines and carbon dioxide by deamination, transamination or decarboxylation. Of course, the α‐ketoic acid produced by the decomposition of amino acids can be converted into sugars, lipids or resynthesized into some nonessential amino acids. It can also be oxidized to carbon dioxide and water through the tricarboxylic acid cycle and release energy. In recent years, there are few reports on the regulation of amino acid metabolism by ncRNAs in HCC.
Glutamine (Gln), a free nonessential amino acid that is abundant in human body, is converted to glutamate by glutaminase and participates in the anabolism of tumor cells. The proliferation of cancer cells absorbs large amounts of glutamine through glutamine transporters and glutamine serves as a major source of energy. Alanine‐serine‐cysteine transporter 2 (ASCT2) is widely expressed in human tissues and organs, and its expression is significantly increased in tumors to meet the rapidly increasing glutamine demand of cancer cells. It was found that miR‐192 and miR‐204 inhibited the expression of lncHOTTIP in HCC through Argonaute 2 (AGO2)‐mediated RNA interference (RNAi), which blocked glutaminolysis (a vital hallmark of cancer cells) of HCC cells and suppress HCC growth by silencing glutaminase1 (GLS1) 143 (Figure 3). Methionine adenosinetransferase (MAT) is a key enzyme in the synthesis of S‐adenosine methionine (SAM), which mainly provides methyl during the metabolism of animal cells. 144 In HCC, upregulation of miR‐495, miR‐664 and miR‐485‐3p can decrease the expression of MAT1A (the gene encoding the catalytic subunit α1 of MAT). After knockout of these miRNAs, cancer cell growth is inhibited and apoptosis is accelerated 145 (Figure 3).
FIGURE 3.
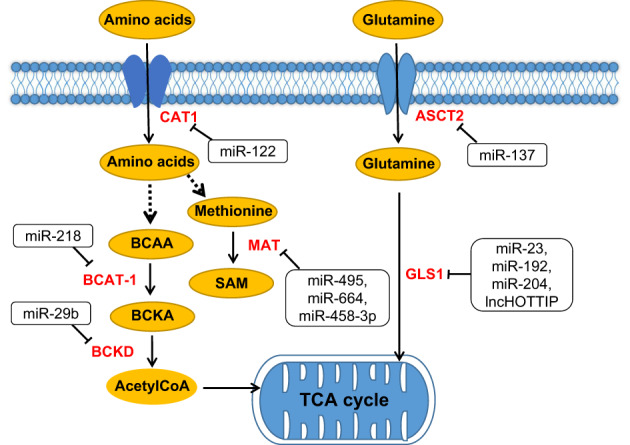
ncRNAs regulate amino acid metabolism in HCC. ncRNAs regulate the metabolism of various amino acids and glutamine (Tables S2 and S3). ASCT2, amino acid transporter 2; BCAA, branched‐chain amino acid; BCAT1, branched‐chain amino acid transaminase 1; BCKA, branched‐chain keto acid; BCKD, branched‐chain ketoacid dehydrogenase; CAT‐1, cationic amino acid transporter 1; GLS1, glutaminase 1; MAT, methionine adenosinetransferase; SAM, sadenosyl‐l‐methionine [Color figure can be viewed at wileyonlinelibrary.com]
3. CONCLUSIONS AND FUTURE PERSPECTIVES
In recent years, the involvement of ncRNAs in the regulation of metabolic reprogramming in cancer have become a research hotspot in the field of biology and oncology. Current evidences suggest that ncRNAs play a significant role in HCC cellular activities. Metabolic reprogramming is considered to be an important hallmark of malignancy. The interaction between ncRNAs and HCC metabolism has attracted extensive attention. Regulation of metabolism of HCC cells by ncRNAs either boosts or inhibits some key enzymes of material metabolism, thus altering the process of HCC progression, such as proliferation, invasion, differentiation and metastasis. Therefore, to some extent, some ncRNAs involved in metabolic regulation could be used as potential biomarkers for HCC.
ncRNAs are extensively involved in the regulation of HCC metabolic reprogramming. Nevertheless, the functions and specific roles of most ncRNAs remain to be studied. On one hand, metabolic reprogramming of HCC provides sufficient energy for tumor growth; on the other hand, it provides a favorable growth environment for tumor progression. Metabolic reprogramming relies on metabolic enzymes, which in turn are regulated by numerous ncRNAs, thus providing many potential therapeutic targets for HCC. Encouragingly, ncRNAs, especially miRNAs, have been extensively and intensively studied in many cancers, including HCC, and have shown promising results in preclinical studies. However, there are still many problems to be solved, such as, what causes the abnormal metabolism? How can we target the metabolic reprogramming of cancer cells to treat HCC? And what is the specific mechanism of metabolic reprogramming in HCC. What's more, most studies on the metabolic reprogramming of ncRNA in HCC are performed without knockout animal models of ncRNA, because no such model has been applied in the study of metabolic reprogramming of HCC so far. Noteworthy, a recent study by Li et al 146 identified two lncRNAs (lncPair and lncHULC) that regulate phenylalanine hydroxylase and enhance phenylalanine metabolism in mice. Besides, in the mouse model of phenylketonuria (lncPair knockout), lncRNA mimics can effectively improve the function of liver phenylalanine hydroxylase and reduce the concentration of phenylalanine in blood. This ncRNA knockout mouse model will help identify key molecules regulating the metabolic reprogramming of HCC and provide theoretical basis for clinical application.
More importantly, understanding the specific mechanism of ncRNA on metabolic reprogramming of HCC will provide a solid theoretical basis for future precision medicine. Although ncRNAs may serve as therapeutic targets for HCC treatment, more research remains needed in the field of ncRNAs.
AUTHOR CONTRIBUTIONS
Yong Zeng, Kefei Yuan and Wenwei Liao conceived the structure of article. Wenwei Liao drafted initial article. Jinpeng Du and Zhen Wang made the figures and tables. Qingbo Feng, Mengheng Liao and Huixian Liu revised the article. All authors read and approved the final article. The work reported in the article has been performed by the authors, unless clearly specified in the text.
CONFLICT OF INTEREST
The authors declare no potential conflict of interests.
Supporting information
Appendix S1 Supporting Information.
Liao W, Du J, Wang Z, et al. The role and mechanism of noncoding RNAs in regulation of metabolic reprogramming in hepatocellular carcinoma. Int J Cancer. 2022;151(3):337‐347. doi: 10.1002/ijc.34040
Wenwei Liao, Jinpeng Du and Zhen Wang contributed equally to this study.
Funding information Natural Science Foundation of China, Grant/Award Numbers: 82070644, 82002572, 82002967, 81972747, 81872004; The National Key Technologies R&D Program, Grant/Award Number: 2018YFC1106800
Contributor Information
Kefei Yuan, Email: yuankefei@scu.edu.cn.
Yong Zeng, Email: zeng_y@scu.edu.cn.
REFERENCES
- 1. Sung H, Ferlay J, Siegel RL, et al. Global cancer statistics 2020: GLOBOCAN estimates of incidence and mortality worldwide for 36 cancers in 185 countries. CA Cancer J Clin. 2021;71(3):209‐249. [DOI] [PubMed] [Google Scholar]
- 2. Wang P, Song X, Cao D, et al. Oncogene‐dependent function of BRG1in hepatocarcinogenesis. Cell Death Dis. 2020;11(2):91. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Warburg O. On the origin of cancer cells. Science. 1956;123:309‐314. [DOI] [PubMed] [Google Scholar]
- 4. Warburg O, Wind F, Negelein E. The metabolism of tumors in the body. J Gen Physiol. 1927;8:519‐530. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Upadhyay M, Samal J, Kandpal M, Singh OV, Vivekanandan P. The Warburg effect: insights from the past decade. Pharmacol Ther. 2013;137:318‐330. [DOI] [PubMed] [Google Scholar]
- 6. Shang RZ, Qu SB, Wang DS. Reprogramming of glucose metabolism in hepatocellular carcinoma: progress and prospects. World J Gastroenterol. 2016;45:9933‐9943. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Calin GA, Croce CM. MicroRNA signatures in human cancers. Nat Rev Cancer. 2006;6:857‐866. [DOI] [PubMed] [Google Scholar]
- 8. Esquela‐Kerscher A, Slack FJ. Oncomirs‐microRNAs with a role in cancer. Nat Rev Cancer. 2006;6:259‐269. [DOI] [PubMed] [Google Scholar]
- 9. Takahashi K, Yan IK, Haga H, Patel T. Modulation of hypoxia‐signaling pathways by extracellular linc‐RoR. J Cell Sci. 2014;127:1585‐1594. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Takahashi K, Yan IK, Kogure T, Haga H, Patel T. Extracellular vesicle‐mediated transfer of long non‐coding RNA ROR modulates chemosensitivity in human hepatocellular cancer. FEBS Open Bio. 2014;4:458‐467. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Zhang Y, Wei W, Cheng N, et al. Hepatitis C virus‐induced up‐regulation of microRNA‐155 promotes hepatocarcinogenesis by activating Wnt signaling. Hepatology. 2012;56(5):1631‐1640. [DOI] [PubMed] [Google Scholar]
- 12. Shen G, Jia H, Tai Q, Li Y, Chen D. MiR‐106b down‐regulates adenomatous polyposis coli and promotes cell proliferation in human hepatocellular carcinoma. Carcinogenesis. 2013;34(1):211‐219. [DOI] [PubMed] [Google Scholar]
- 13. Le MT, Teh C, Shyh‐Chang N, et al. MicroRNA‐125b is a novel negative regulator of p53. Genes Dev. 2009;23:862‐876. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Hu W, Chan CS, Wu R, et al. Negative regulation of tumor suppressor p53 by microRNA miR‐504. Mol Cell. 2010;38:689‐699. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Tang H, Li RP, Liang P, Zhou YL, Wang GW. miR‐125a inhibits the migration and invasion of liver cancer cells via suppression of the PI3K/AKT/mTOR signaling pathway. Oncol Lett. 2015;10:681‐686. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. He Y, Meng XM, Huang C, et al. Long noncoding RNAs: novel insights into hepatocelluar carcinoma. Cancer Lett. 2014;344(1):20‐27. [DOI] [PubMed] [Google Scholar]
- 17. Ma X, Mo M, Tan C, et al. Liver‐specific LINC01146, a promising prognostic indicator, inhibits the malignant phenotype of hepatocellular carcinoma cells both in vitro and in vivo. J Transl Med. 2022;20(1):1‐18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Liu Y, Geng X. Long non‐coding RNA (lncRNA) CYTOR promotes hepatocellular carcinoma proliferation by targeting the microRNA‐125a‐5p/LASP1 axis. Bioengineered. 2022;13(2):3666‐3679. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Matouk IJ, Mezan S, Mizrahi A, et al. The oncofetal H19 RNA connection: hypoxia, p53 and cancer. Biochem Biophys Acta. 2010;1803:443‐451. [DOI] [PubMed] [Google Scholar]
- 20. Yuan JH, Yang F, Wang F, et al. A long non‐coding RNA activated by TGF‐beta promotes the invasion‐metastasis cascade in hepatocellular carcinoma. Cancer Cell. 2014;25(5):666‐681. [DOI] [PubMed] [Google Scholar]
- 21. Wang L, Long H, Zheng Q, Bo X, Xiao X, Li B. Circular RNA circRHOT1 promotes hepatocellular carcinoma progression by initiation of NR2F6 expression. Mol Cancer. 2019;18(1):119. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Yu J, Xu QG, Wang ZG, et al. Circular RNA cSMARCA5 inhibits growth and metastasis in hepatocellular carcinoma. J Hepatol. 2018;68:1214‐1227. [DOI] [PubMed] [Google Scholar]
- 23. Li P, Song R, Yin F, et al. circMRPS35 promotes malignant progression and cisplatin resistance in hepatocellular carcinoma. Mol Ther. 2022;30(1):431‐447. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Liu D, Liu W, Chen X, et al. circKCNN2 suppresses the recurrence of hepatocellular carcinoma at least partially via regulating miR‐520c‐3p/methyl‐DNA‐binding domain protein 2 axis. Clin Transl Med. 2022;12(1):e662. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Zhuang X, Chen Y, Wu Z, et al. Mitochondrial miR‐181a‐5p promotes glucose metabolism reprogramming in liver cancer by regulating the electron transport chain. Carcinogenesis. 2020;41(7):972‐983. [DOI] [PubMed] [Google Scholar]
- 26. Geschwind JF, Georgiades CS, Ko YH, Pedersen PL. Recently elucidated energy catabolism pathways provide opportunities for novel treatments in hepatocellular carcinoma. Expert Rev Anticancer Ther. 2004;4:449‐457. [DOI] [PubMed] [Google Scholar]
- 27. Ko YH, Verhoeven HA, Lee MJ, Corbin DJ, Vogl TJ, Pedersen PL. A translational study “case report” on the small molecule “energy blocker” 3‐bromopyruvate (3BP) as a potent anticancer agent: from bench side to bedside. J Bioenerg Biomembr. 2012;44:163‐170. [DOI] [PubMed] [Google Scholar]
- 28. Mattick JS. Challenging the dogma: the hidden layer of non‐protein‐coding RNAs in complex organisms. Bioessays. 2003;25:930‐939. [DOI] [PubMed] [Google Scholar]
- 29. Mattick JS. The hidden genetic program of complex organisms. Sci Am. 2004;291:60‐67. [DOI] [PubMed] [Google Scholar]
- 30. Hombach S, Kretz M. Non‐coding RNAs: classification, biology and functioning. Adv Exp Med Biol. 2016;937:3‐17. [DOI] [PubMed] [Google Scholar]
- 31. Heinrichs A. MicroRNAs get a boost. Nat Rev Mol Cell Biol. 2009;10(5):302‐303. [DOI] [PubMed] [Google Scholar]
- 32. Baumann K. Gene expression: RNAi as a global transcriptional activator. Nat Rev Mol Cell Biol. 2014;15:298. [DOI] [PubMed] [Google Scholar]
- 33. Sato K, Siomi MC. Piwi‐interacting RNAs: biological functions and biogenesis. Essays Biochem. 2013;54:39‐52. [DOI] [PubMed] [Google Scholar]
- 34. Ross RJ, Weiner MM, Lin HF. PIWI proteins and PIWI‐interacting RNAs in the soma. Nature. 2014;505:353‐359. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Kim VN, Han J, Siomi MC. Biogenesis of small RNAs in animals. Nat Rev Mol Cell Biol. 2009;10:126‐139. [DOI] [PubMed] [Google Scholar]
- 36. Bonasio R, Shiekhattar R. Regulation of transcription by long non‐coding RNAs. Annu Rev Genet. 2014;48:433‐455. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Brown JW, Marshall DF, Echeverria M. Intronic noncoding RNAs and splicing. Trends Plant Sci. 2008;13(7):335‐342. [DOI] [PubMed] [Google Scholar]
- 38. Shankaraiah RC, Veronese A, Sabbioni S, Negrini M. Non‐coding RNAs in the reprogramming of glucose metabolism in cancer. Cancer Lett. 2018;419:167‐174. [DOI] [PubMed] [Google Scholar]
- 39. Wong CM, Tsang HC, Ng OL. Non‐coding RNAs in hepatocellular carcinoma: molecular functions and pathological implications. Nat Rev Gastroenterol Hepatol. 2018;15(3):137‐151. [DOI] [PubMed] [Google Scholar]
- 40. Fu XD. Non‐coding RNA: a new frontier in regulatory biology. Natl Sci Rev. 2014;1:190‐204. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Kugel JF, Goodrich JA. Non‐coding RNAs: key regulators of mammalian transcription. Trends Biochem Sci. 2012;37:144‐151. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Mondal T, Kanduri C. Maintenance of epigenetic information: a non‐coding RNA perspective. Chromosome Res. 2013;21:615‐625. [DOI] [PubMed] [Google Scholar]
- 43. Chan B, Manley J, Lee J, Singh SR. The emerging roles of microRNAs in cancer metabolism. Cancer Lett. 2015;356:301‐308. [DOI] [PubMed] [Google Scholar]
- 44. Pulito C, Donzelli S, Muti P, et al. MicroRNAs and cancer metabolism reprogramming: the paradigm of metformin. Ann Transl Med. 2014;2:58. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Zhao XY, Lin JD. Long non‐coding RNAs: a new regulatory code in metabolic control. Trends Biochem Sci. 2015;40:586‐596. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Sanger HL, Klotz G, Riesner D, Gross HJ, Kleinschmidt AK. Viroids are single‐stranded covalently closed circular RNA molecules existing as highly base‐paired rod‐like structures. Proc Natl Acad Sci U S A. 1976;73:3852‐3856. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Salzman J. Circular RNA expression: its potential regulation and function. Trends Genet. 2016;32:309‐316. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48. Jeck WR, Sharpless NE. Detecting and characterizing circular RNAs. Nat Biotechnol. 2014;32:453‐461. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49. Hansen TB, Jensen TI, Clausen BH, et al. Natural RNA circles function as efficient microRNA sponges. Nature. 2013;495:384‐388. [DOI] [PubMed] [Google Scholar]
- 50. Memczak S, Jens M, Elefsinioti A, et al. Circular RNAs are a large class of animal RNAs with regulatory potency. Nature. 2013;495:333‐338. [DOI] [PubMed] [Google Scholar]
- 51. Vo JN, Cieslik M, Zhang Y, et al. The landscape of circular RNA in cancer. Cell. 2019;176(4):869‐881. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52. Glažar P, Papavasileiou P, Rajewsky N. circBase: a database for circular RNAs. RNA. 2014;20(11):1666‐1670. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53. Piwecka M, Glažar P, Hernandez‐Miranda LR, et al. Loss of a mammalian circular RNA locus causes miRNA deregulation and affects brain function. Science. 2017;357(6357):1254. [DOI] [PubMed] [Google Scholar]
- 54. Zheng Q, Bao C, Guo W, et al. Circular RNA profiling reveals an abundant circHIPK3 that regulates cell growth by sponging multiple miRNAs. Nat Commun. 2016;7(1):1‐13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55. Yu L, Gong X, Sun L, Zhou Q, Lu B, Zhu L. The circular RNA Cdr1as act as an oncogene in hepatocellular carcinoma through targeting miR‐7 expression. PLoS One. 2016;11(7):e0158347. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 56. Gong Y, Mao J, Wu DI, et al. 33 promotes the proliferation of human HCC by sponging miR‐200a‐3p and upregulating CDK6. Cancer Cell Int. 2018;18(1):1‐9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57. Xu L, Feng X, Hao X, et al. CircSETD3 (Hsa_circ_0000567) acts as a sponge for microRNA‐421 inhibiting hepatocellular carcinoma growth. J Exp Clin Cancer Res. 2019;38(1):1‐15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58. Han D, Li J, Wang H, et al. Circular RNA circMTO1 acts as the sponge of microRNA‐9 to suppress hepatocellular carcinoma progression. Hepatology. 2017;66(4):1151‐1164. [DOI] [PubMed] [Google Scholar]
- 59. Zhang X, Luo P, Jing W, Zhou H, Liang C, Tu J. circSMAD2 inhibits the epithelial–mesenchymal transition by targeting miR‐629 in hepatocellular carcinoma. OncoTargets Ther. 2018;11:2853‐2863. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60. Ning B, Peng E, Qiu X, et al. circFBLIM1 act as a ceRNA to promote hepatocellular cancer progression by sponging miR‐346. J Exp Clin Cancer Res. 2018;37(1):1‐9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61. Chen G, Shi Y, Liu M, Sun J. circHIPK3 regulates cell proliferation and migration by sponging miR‐124 and regulating AQP3 expression in hepatocellular carcinoma. Cell Death Dis. 2018;9(2):1‐13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62. Kristensen LS, Hansen TB, Venø MT, Kjems J. Circular RNAs in cancer: opportunities and challenges in the field. Oncogene. 2018;37(5):555‐565. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63. Li Q, Pan X, Zhu D, Deng Z, Jiang R, Wang X. Circular RNA MAT2B promotes glycolysis and malignancy of hepatocellular carcinoma through the miR‐338‐3p/PKM2 axis under hypoxic stress. Hepatology. 2019;70(4):1298‐1316. [DOI] [PubMed] [Google Scholar]
- 64. Zhang B, Liu Z, Cao K, et al. Circ‐SPECC1 modulates TGFβ2 and autophagy under oxidative stress by sponging miR‐33a to promote hepatocellular carcinoma tumorigenesis. Cancer Med. 2020;9(16):5999‐6008. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 65. Zhong L, Wang Y, Cheng Y, et al. Circular RNA circC3P1 suppresses hepatocellular carcinoma growth and metastasis through miR‐4641/PCK1 pathway. Biochem Biophys Res Commun. 2018;499:1044‐1049. [DOI] [PubMed] [Google Scholar]
- 66. Li J, Hu ZQ, Yu S, et al. CircRPN2 inhibits aerobic glycolysis and metastasis in hepatocellular carcinoma. Cancer Res. 2022;82:1055‐1069. [DOI] [PubMed] [Google Scholar]
- 67. Esteller M. Non‐coding RNAs in human disease. Nat Rev Genet. 2011;12(12):861‐874. [DOI] [PubMed] [Google Scholar]
- 68. Williams GT, Farzaneh F. Are snoRNAs and snoRNA host genes new players in cancer? Nat Rev Cancer. 2012;12(2):84‐88. [DOI] [PubMed] [Google Scholar]
- 69. Mannoor K, Liao JP, Jiang F. Small nucleolar RNAs in cancer. Biochim Biophys Acta. 2012;1826:121‐128. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70. Stepanov GA, Filippova JA, Komissarov AB, Kuligina EV, Richter VA, Semenov DV. Regulatory role of small nucleolar RNAs in human diseases. Biomed Res Int. 2015;2015:1‐10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71. Watkins NJ, Bohnsack MT. The box C/D and H/ACA snoRNPs: key players in the modifi cation, processing and the dynamic folding of ribosomal RNA. Wiley Interdiscip Rev RNA. 2012;3:397‐414. [DOI] [PubMed] [Google Scholar]
- 72. Yu YT, Meier UT. RNA‐guided isomerization of uridine to pseudouridine‐pseudouridylation. RNA Biol. 2014;11:1483‐1494. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73. Farazi TA, Juranek SA, Tuschl T. The growing catalog of small RNAs and their association with distinct Argonaute/Piwi family members. Development. 2008;135:1201‐1214. [DOI] [PubMed] [Google Scholar]
- 74. Luteijn MJ, Ketting RF. PIWI‐interacting RNAs: from generation to transgenerational epigenetics. Nat Rev Genet. 2013;14:523‐534. [DOI] [PubMed] [Google Scholar]
- 75. Siomi MC, Sato K, Pezic D, Aravin AA. PIWIinteracting small RNAs: the vanguard of genome defence. Nat Rev Mol Cell Biol. 2011;12:246‐258. [DOI] [PubMed] [Google Scholar]
- 76. Bartel DP. MicroRNAs: target recognition and regulatory functions. Cell. 2009;136:215‐233. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77. Cannell IG, Kong YW, Bushell M. How do microRNAs regulate gene expression? Biochem Soc Trans. 2008;36:1224‐1231. [DOI] [PubMed] [Google Scholar]
- 78. Lim LP, Lau NC, Garrett‐Engele P, et al. Microarray analysis shows that some microRNAs downregulate large numbers of target mRNAs. Nature. 2005;433(7027):769‐773. [DOI] [PubMed] [Google Scholar]
- 79. Vander Heiden MG, Cantley LC, Thompson CB. Understanding the Warburg efect: the metabolic requirements of cell proliferation. Science. 2009;324(5930):1029‐1033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80. Lunt SY, Heiden MV. Aerobic glycolysis: meeting the metabolic requirements of cell proliferation. Annu Rev Cell Dev Biol. 2011;27(1):441‐464. [DOI] [PubMed] [Google Scholar]
- 81. Hu S, Balakrishnan A, Bok RA, et al. 13C‐pyruvate imaging reveals alterations in glycolysis that precede c‐Myc‐induced tumor formation and regression. Cell Metab. 2011;14(1):131‐142. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82. Gao P, Sun L, He X, Cao Y, Zhang H. MicroRNAs and the Warburg effect: new players in an old arena. Curr Gene Ther. 2012;12(4):285‐291. [DOI] [PubMed] [Google Scholar]
- 83. Rottiers V, Näär AM. MicroRNAs in metabolism and metabolic disorders. Nat Rev Mol Cell Biol. 2012;13(4):239‐250. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84. Zhang LF, Lou JT, Lu MH, et al. Suppression of miR‐199a maturation by HuR iscrucial for hypoxia‐induced glycolytic switch inhepatocellular carcinoma. EMBO J. 2015;34(21):2671‐2685. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85. Zhang LF, Jiang S, Liu MF. MicroRNA regulation and analytical methods in cancer cell metabolism. Cell Mol Life Sci. 2017;74(16):2929‐2941. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86. Sun L, He X, Cao Y, et al. MicroRNAs and energy metabolism in cancer cells. In: Babashah S, ed. MicroRNAs: Key Regulators of Oncogenesis. Cham: Springer; 2014:83‐95. [Google Scholar]
- 87. Xu Q, Zhang M, Tu J, Pang L, Cai W, Liu X. MicroRNA‐122 affects cell aggressiveness and apoptosis by targeting PKM2 in human hepatocellular carcinoma. Oncol Rep. 2015;34(4):2054‐2064. [DOI] [PubMed] [Google Scholar]
- 88. Liu AM, Xu Z, Shek FH, et al. miR‐122 targets pyruvate kinase M2 and affects metabolism of hepatocellular carcinoma. PLoS One. 2014;9(1):e86872. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89. Fong MY, Zhou W, Liu L, et al. Breast‐cancer‐secreted miR‐122 reprograms glucose metabolism in premetastatic niche to promote metastasis. Nat Cell Biol. 2015;17:183‐194. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90. Wu H, Wang Y, Wu C, Yang P, Li H, Li Z. Resveratrol induces cancer cell apoptosis through miR‐326/PKM2‐ mediated ER stress and mitochondrial fission. J Agric Food Chem. 2016;64:9356‐9367. [DOI] [PubMed] [Google Scholar]
- 91. Jin F, Wang Y, Zhu Y, et al. The miR‐125a/HK2 axis regulates cancer cell energy metabolism reprogramming in hepatocellular carcinoma. Sci Rep. 2017;7(1):3089. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92. Wang J, Chen J, Sun F, et al. miR‐202 functions as a tumor suppressor in hepatocellular carcinoma by targeting HK2. Oncol Lett. 2020;19(3):2265‐2271. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93. Xu F, Yan JJ, Gan Y, et al. miR‐885‐5p negatively regulates Warburg effect by silencing hexokinase 2 in liver cancer. Mol Ther Nucleic Acids. 2019;18:308‐319. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94. Ramirez CM, Goedeke L, Rotllan N, et al. MicroRNA 33 regulates glucose metabolism. Mol Cell Biol. 2013;33:2891‐2902. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95. Hermeking H. MicroRNAs in the p53 network: micromanagement of tumour suppression. Nat Rev Cancer. 2012;12:613‐626. [DOI] [PubMed] [Google Scholar]
- 96. Lynn FC. Meta‐regulation: MicroRNA regulation of glucose and lipid metabolism. Trends Endocrinol Metab. 2009;20:452‐459. [DOI] [PubMed] [Google Scholar]
- 97. Lin Y, Wu M, Huang Y, et al. Taurine upregulated gene 1 functions as a master regulator to coordinate glycolysis and metastasis in hepatocellular carcinoma. Hepatology. 2018;67:188‐203. [DOI] [PubMed] [Google Scholar]
- 98. Subramaniam S, Jeet V, Clements JA, Gunter JH, Batra J. Emergence of microRNAs as key players in cancer cell metabolism. Clin Chem. 2019;65(9):1090‐1101. [DOI] [PubMed] [Google Scholar]
- 99. Hatziapostolou M, Polytarchou C, Iliopoulos D. miRNAs link metabolic reprogramming to oncogenesis. Trends Endocrinol Metab. 2013;24:361‐373. [DOI] [PubMed] [Google Scholar]
- 100. Liu LZ, Li C, Chen Q, et al. miR‐21 induced angiogenesis through AKT and ERK activation and HIF‐1α expression. PLoS One. 2011;6:e19139. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101. Cai Y, He T, Liang L, Zhang X, Yuan H. Upregulation of microRNA‐337 promotes the proliferation of endometrial carcinoma cells via targeting PTEN. Mol Med Rep. 2016;13(6):4827‐4834. [DOI] [PubMed] [Google Scholar]
- 102. Chen J, Yan D, Wu W, et al. MicroRNA‐130a promotes the metastasis and epithelial‐mesenchymal transition of osteosarcoma by targeting PTEN. Oncol Rep. 2016;35(6):3285‐3292. [DOI] [PubMed] [Google Scholar]
- 103. Wang B, Sun F, Dong N, et al. MicroRNA‐7 directly targets insulin‐like growth factor 1 receptor to inhibit cellular growth and glucose metabolism in gliomas. Diagn Pathol. 2014;9:211. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 104. Sayed D, Abdellatif M. AKT‐ing via microRNA. Cell Cycle. 2010;9(16):3233‐3237. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 105. Brandon F, Ashley S, Deberardinis RJ. Metabolic reprogramming and cancer progression. Science. 2020;368:368. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 106. Liu W, Kang L, Han J, et al. miR‐342‐3p suppresses hepatocellular carcinoma proliferation through inhibition of IGF‐1R‐mediated Warburg effect. Onco Targets Ther. 2018;11:1643‐1653. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 107. Hu Y, Yang Z, Bao D, Ni J, Lou J. miR‐455‐5p suppresses hepatocellular carcinoma cell growth and invasion via IGF‐1R/AKT/GLUT1 pathway by targeting IGF‐1R. Pathol Res Pract. 2019;215:152674. [DOI] [PubMed] [Google Scholar]
- 108. Chen Z, Zuo X, Zhang Y, et al. MiR‐3662 suppresses hepatocellular carcinoma growth through inhibition of HIF‐1α‐mediated Warburg effect. Cell Death Dis. 2018;9(5):1‐14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 109. Okazaki Y, Furuno M, Kasukawa T, et al. Analysis of the mouse transcriptome based on functional annotation of 60,770 full‐length cDNAs. Nature. 2002;420:563‐573. [DOI] [PubMed] [Google Scholar]
- 110. Haemmerle M, Gutschner T. Long non‐coding RNAs in cancer and development: where do we go from here? Int J Mol Sci. 2015;16:1395‐1405. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 111. Karapetyan AR, Buiting C, Kuiper RA, Coolen M. Regulatory roles for long ncRNA and mRNA. Cancer. 2013;5:462‐490. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 112. Prensner JR, Chinnaiyan AM. The emergence of lncRNAs in cancer biology. Cancer Discov. 2011;1:391‐407. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 113. Zheng YL, Li L, Jia YX, et al. LINC01554‐mediated glucose metabolism reprogramming suppresses tumorigenicity in hepatocellular carcinoma via downregulating PKM2 expression and inhibiting Akt/mTOR signaling pathway. Theranostics. 2019;9:796‐810. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 114. Yang F, Zhang H, Mei Y, Wu M. Reciprocal regulation of HIF‐1α and lincRNA‐p21 modulates the Warburg effect. Mol Cell. 2014;53:88‐100. [DOI] [PubMed] [Google Scholar]
- 115. Cheng L, Hu S, Ma J, et al. Long noncoding RNA RP11‐241J12.3 targeting pyruvate carboxylase promotes hepatocellular carcinoma aggressiveness by disrupting pyruvate metabolism and the DNA mismatch repair system. Mol Biomed. 2022;3(1):1‐16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 116. Wang W, Deng Z, Jin Z, et al. Bioinformatics analysis and experimental verification of five metabolism‐related lncRNAs as prognostic models for hepatocellular carcinoma. Medicine. 2022;101(4):e28694. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 117. Yuan SX, Yang F, Yang Y, et al. Long non‐coding RNA associated with microvascular invasion in hepatocellular carcinoma promotes angiogenesis and serves as a predictor for hepatocellular carcinoma patients' poor recurrence‐free survival after hepatectomy. Hepatology. 2012;56(6):2231‐2241. [DOI] [PubMed] [Google Scholar]
- 118. Huang JL, Zheng L, Hu YW, Wang Q. Characteristics of long non‐coding RNA and its relation to hepatocellular carcinoma. Carcinogenesis. 2014;35(3):507‐514. [DOI] [PubMed] [Google Scholar]
- 119. Malakar P, Stein I, Saragovi A, et al. Long noncoding RNA MALAT1 regulates cancer glucose metabolism by enhancing mTOR‐mediated translation of TCF7L2. Cancer Res. 2019;79(10):2480‐2493. [DOI] [PubMed] [Google Scholar]
- 120. Law PTY, Qin H, Ching AKK, et al. Deep sequencing of small RNA transcriptome reveals novel non‐coding RNAs in hepatocellular carcinoma. J Hepatol. 2013;58(6):1165‐1173. [DOI] [PubMed] [Google Scholar]
- 121. Xu G, Yang F, Ding CL, et al. Small nucleolar RNA 113‐1 suppresses tumorigenesis in hepatocellular carcinoma. Mol Cancer. 2014;13(1):1‐14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 122. Fang X, Yang D, Luo H, et al. SNORD126 promotes HCC and CRC cell growth by activating the PI3K‐AKT pathway through FGFR2. J Mol Cell Biol. 2017;9(3):243‐255. [DOI] [PubMed] [Google Scholar]
- 123. Fernandez‐Hernando C, Suarez Y, Rayner KJ, et al. MicroRNAs in lipid metabolism. Curr Opin Lipidol. 2011;22:86‐92. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 124. Santos CR, Schulze A. Lipid metabolism in cancer. FEBS J. 2012;279:2610‐2623. [DOI] [PubMed] [Google Scholar]
- 125. Currie E, Schulze A, Zechner R, Walther TC, Farese RV Jr. Cellular fatty acid metabolism and cancer. Cell Metab. 2013;18:153‐161. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 126. Krutzfeldt J, Rajewsky N, Braich R, et al. Silencing of microRNAs in vivo with ‘antagomirs’. Nature. 2005;438:685‐689. [DOI] [PubMed] [Google Scholar]
- 127. Rotllan N, Fernandez‐Hernando C. MicroRNA regulation of cholesterol metabolism. Cholesterol. 2012;2012:1‐8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 128. Esau C, Davis S, Murray SF, et al. miR‐122 regulation of lipid metabolism revealed by in vivo antisense targeting. Cell Metab. 2006;3(2):87‐98. [DOI] [PubMed] [Google Scholar]
- 129. Davalos A, Goedeke L, Smibert P, et al. MiR‐33a/b contribute to the regulation of fatty acid metabolism and insulin signaling. Proc Natl Acad Sci U S A. 2011;108:9232‐9237. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 130. Jeon TI, Osborne TF. SREBPs: metabolic integrators in physiology and metabolism. Trends Endocrinol Metab. 2012;23:65‐72. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 131. Shao W, Espenshade PJ. Expanding roles for SREBP in metabolism. Cell Metab. 2012;16:414‐419. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 132. Wen YA, Xiong X, Zaytseva YY, et al. Downregulation of SREBP inhibits tumor growth and initiation by altering cellular metabolism in colon cancer. Cell Death Dis. 2018;9:265. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 133. Najafi‐Shoushtari SH, Kristo F, Li Y, et al. MicroRNA‐33 and the SREBP host genes cooperate to control cholesterol homeostasis. Science. 2010;328:1566‐1569. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 134. Rayner KJ, Suarez Y, Davalos A, et al. MiR‐33 contributes to the regulation of cholesterol homeostasis. Science. 2010;328:1570‐1573. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 135. Niesor EJ, Schwartz GG, Perez A, et al. Statin‐induced decrease in ATP‐binding cassette transporter A1 expression via microRNA33 induction may counteract cholesterol efflux to high‐density lipoprotein. Cardiovasc Drugs Ther. 2015;29:7‐14. [DOI] [PubMed] [Google Scholar]
- 136. Lee J, Hong SW, Park SE, et al. AMP‐activated protein kinase suppresses the expression of LXR/SREBP‐1 signaling‐induced ANGPTL8 in HepG2 cells. Mol Cell Endocrinol. 2015;414:148‐155. [DOI] [PubMed] [Google Scholar]
- 137. Fernandez‐Hernando C, Ramirez CM, Goedeke L, et al. MicroRNAs in metabolic disease. Arterioscler Thromb Vasc Biol. 2013;33:178‐185. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 138. Zheng Y, Jiang S, Zhang Y, Zhang R, Gong D. Detection of miR‐33 expression and the verification of its target genes in the fatty liver of geese. Int J Mol Sci. 2015;16:12737‐12752. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 139. Panzitt K, Tschernatsch MM, Guelly C, Moustafa T, et al. Characterization of HULC, a novel gene with striking upregulation in hepatocellular carcinoma, as noncoding RNA. Gastroenterology. 2007;132(1):330‐342. [DOI] [PubMed] [Google Scholar]
- 140. Hammerle M, Gutschner T, Uckelmann H, et al. Posttranscriptional destabilization of the liver‐specific long non‐coding RNA HULC by the IGF2 mRNA‐binding protein 1 (IGF2BP1). Hepatology. 2013;58:1703‐1712. [DOI] [PubMed] [Google Scholar]
- 141. Cui M, Xiao Z, Wang Y, et al. Long non‐coding RNA HULC modulates abnormal lipid metabolism in hepatoma cells through an miR‐9‐mediated RXRA signaling pathway. Cancer Res. 2015;75(5):846‐857. [DOI] [PubMed] [Google Scholar]
- 142. Søndergaard JN, Sommerauer C, Atanasoai I, et al. CCT3‐LINC00326 axis regulates hepatocarcinogenic lipid metabolism. Gut. 2022;12(1):325109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 143. Ge Y, Yan X, Jin Y, et al. MiRNA‐192 and miRNA‐204 directly suppress lncRNA HOTTIP and interrupt GLS1‐mediated glutaminolysis in hepatocellular carcinoma. PLoS Genet. 2015;11(12):e1005726. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 144. Lu SC, Mato JM. S‐Adenosylmethionine in cell growth, apoptosis and liver cancer. J Gastroenterol Hepatol. 2008;23:S73‐S77. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 145. Yang HP, Cho ME, Li T, et al. MicroRNAs regulate methionine adenosyltransferase 1A expression in hepatocellular carcinoma. J Clin Investig. 2013;123:285‐298. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 146. Li Y, Tan Z, Zhang Y, et al. A noncoding RNA modulator potentiates phenylalanine metabolism in mice. Science. 2021;373(6555):662‐673. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Appendix S1 Supporting Information.


