Figure 1.
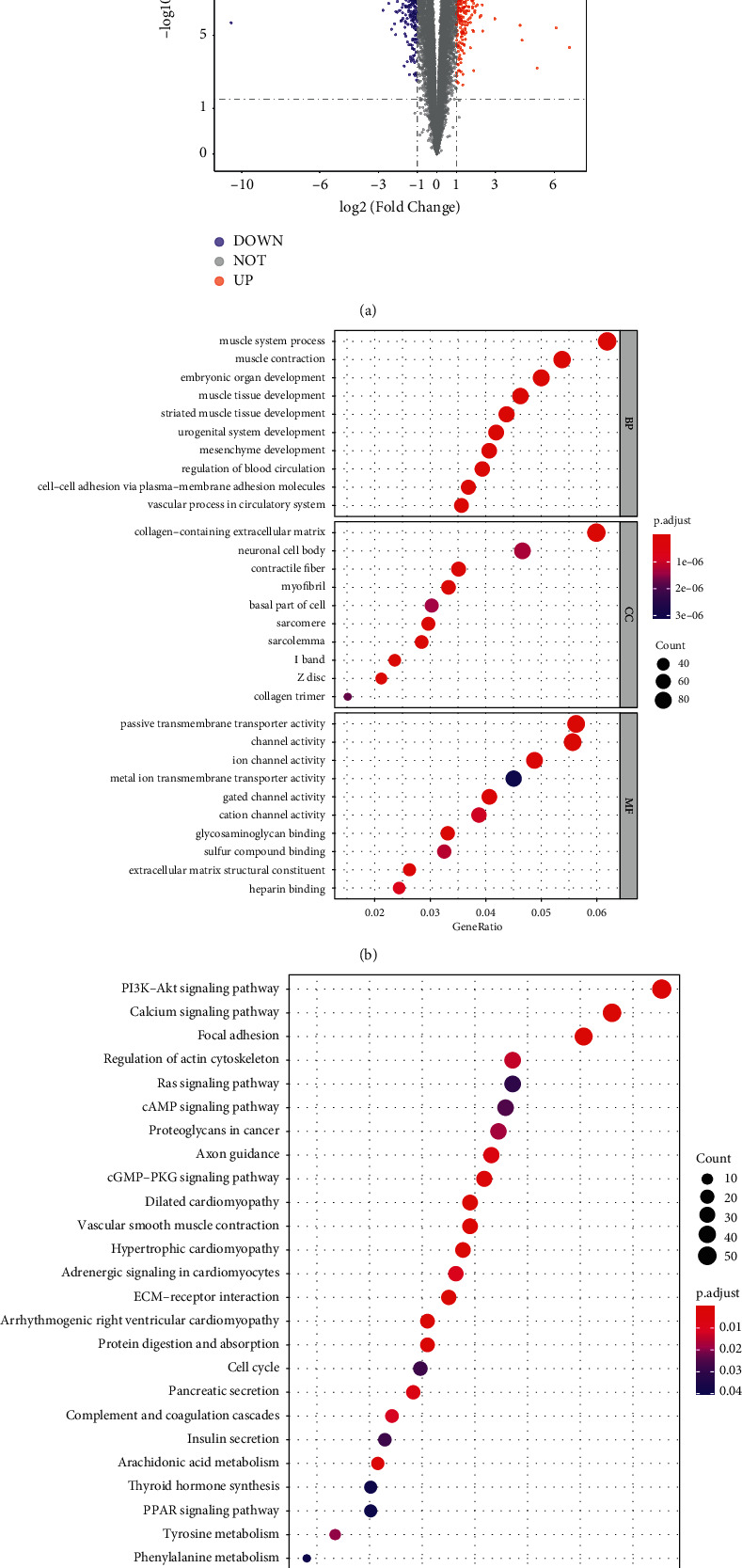
Multiple functions of DEGs have been identified in the cancer genome atlas prostate cancer (TCGA-PRAD) cohort. (a) DEGs between PRAD and control samples were identified using fold-change values and adjusted p-values. DEGs, differentially expressed genes; PRAD, prostate cancer; Control, normal tissue. Red and blue dots indicate upregulated and down-regulated mRNAs, respectively. Statistical significance was set at p-value <0.05. (b). DEGs enriched in the gene ontology (GO) pathways. BP, biological process; CC, cellular component; MF, molecular function. Adjusted p value <0.05 was considered statistically significant. (c) DEGs enriched in the Kyoto Encyclopedia of Genes and Genomes (KEGG) pathways. Adjusted p value <0.05 was considered statistically significant.
