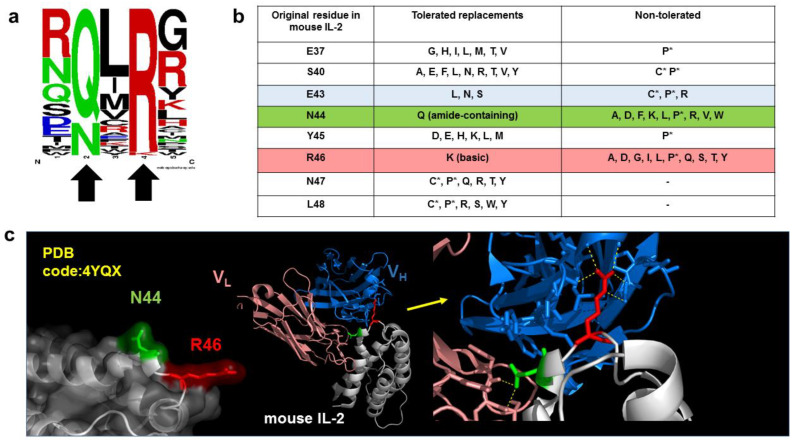
Figure 3.
Identification of an epitope in mouse Interleukin-2 by different methods (a) The weblogo represents the common peptide motif identified in 45 out of 47 peptide ligands selected from a random phage-displayed decapeptide library with the antibody JES6-1A12 (anti-mouse Interleukin-2). The recurrent motif (Q/N)X(R/K) is distinguished by the presence of an amide-containing residue (Q/N, represented in green) and a basic amino acid (R, occasionally replaced by K, red), which are indicated with arrows [11]. The residue between them, as well as the flanking amino acids (aa), are highly variable. Letter size is proportional to the frequency of a given residue among selected peptides. (b) Mutagenesis scanning of phage-displayed mouse IL-2 with the same antibody revealed the importance of N44 and R46, which could only be replaced by the other amide-containing and basic amino acid, respectively [11]. E43 is a minor contributor, as shown by the existence of tolerated and nontolerated mutations at this position. The other residues could be substituted by different aa with diverse properties without affecting antibody recognition. In some cases, replacements by Cys and Pro were not tolerated, probably due to their effects on global protein structure. (c) Crystal structure of the antigenic region that contains N44 and R46 (left), and its interface with JES6-1A12 variable domains (center) [77]. The zoom-in view (right) shows the interactions of the two already identified critical aa (N44 and R46) with light and heavy chain variable regions. Dotted yellow lines indicate polar contacts established by them.