Figure 5. Residues necessary for Spo0A dimerization in other Firmicutes are functionally conserved in C. difficile.
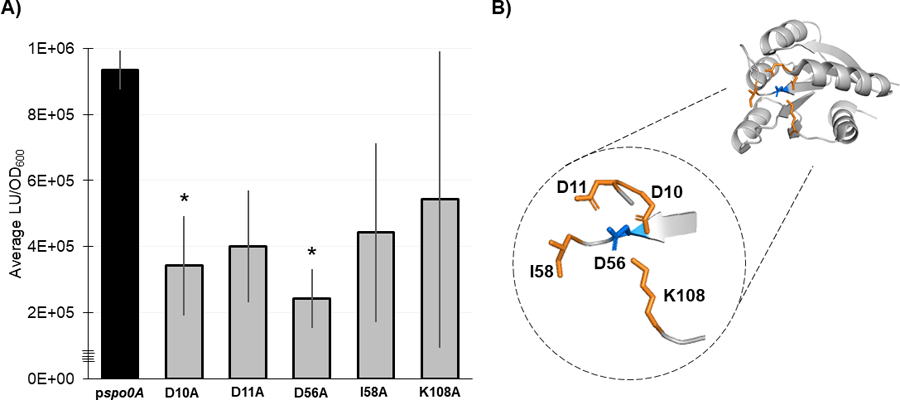
A) Split-luciferase activity in strains 630Δerm spo0A pspo0A (MC1906) and the Spo0A site-directed mutants D10A (MC2001), D11A (MC2002), D56A (MC2003), I58A (MC2004), and K108A (MC2005) fused to SmBit and LgBit fragments after cultures were grown in 70:30 sporulation broth to OD600 = 0.8 – 0.9 and induced with anhydrous tetracycline (ATc) for 1 h. Average luminescence outputs are normalized to optical densities (LU/OD600). Error bars represent the standard deviation of three independent experiments (*, P = < 0.05) as determined by a one-way ANOVA with Dunnett’s multiple comparisons test. B) 3D structure of Spo0A with the residues that form the aspartyl pocket and facilitate dimerization highlighted orange near the site of activation (D56, highlighted blue). Spo0A PDB code 5WQ0, edited in PyMOL (The PyMOL Molecular Graphics System, Version 2.4.0 Schrödinger, LLC).
