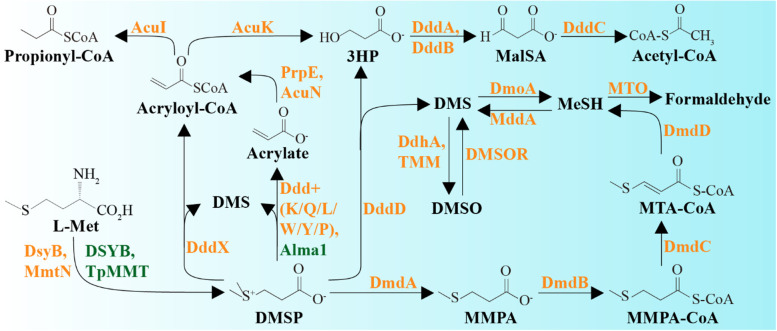
Fig. 1.
Pathways of DMSP synthesis and degradation. DMSP can be synthesised by both phytoplankton and bacterioplankton from methionine (L-Met). The SAM-dependent S-methyltransferase of the transamination pathway has been identified from phytoplankton (DSYB and TpMMT), and bacterioplankton (DsyB). Bacteria also can synthesise DMSP through a methylation pathway mediated by a L-Met-S-methylating enzyme MmtN. DMSP can be degraded through two competing pathways. The demethylation pathway involves DmdABCD and leads to the formation of methylmercaptopropionate (MMPA), methylthioacryloyl-CoA (MTA-CoA) and methanethiol (MeSH). MeSH can be oxidised to formaldehyde by MeSH oxidase (MTO). The cleavage pathway catalysed by Ddd enzymes in some bacteria, fungi and viruses, or Alma1 in algae, liberates DMS and acrylate, acryloyl-CoA or 3-hydroxypropionate (3HP). DMS can be further oxidised by marine microbes through trimethylamine monooxygenase (TMM) or dimethylsulfide dehydrogenase (DdhA) to generate dimethyl sulfoxide or by dimethylsulfide monooxgenase (DmoA) to generate MeSH. Methanethiol S-methyltransferase (MddA) and dimethyl sulfoxide reductase (DMSOR) mediate the production of DMS from MeSH and DMSO, respectively. Potentially toxic acrylate is detoxified by bacteria through several enzymes (PrpE, AcuI, AcuN and AcuK). Bacterial catabolism of 3HP involves DddABC proteins and generates malonate semi-aldehyde (MalSA) and acetyl-CoA. Enzymes from phytoplankton and bacteria are shown in green and orange, respectively