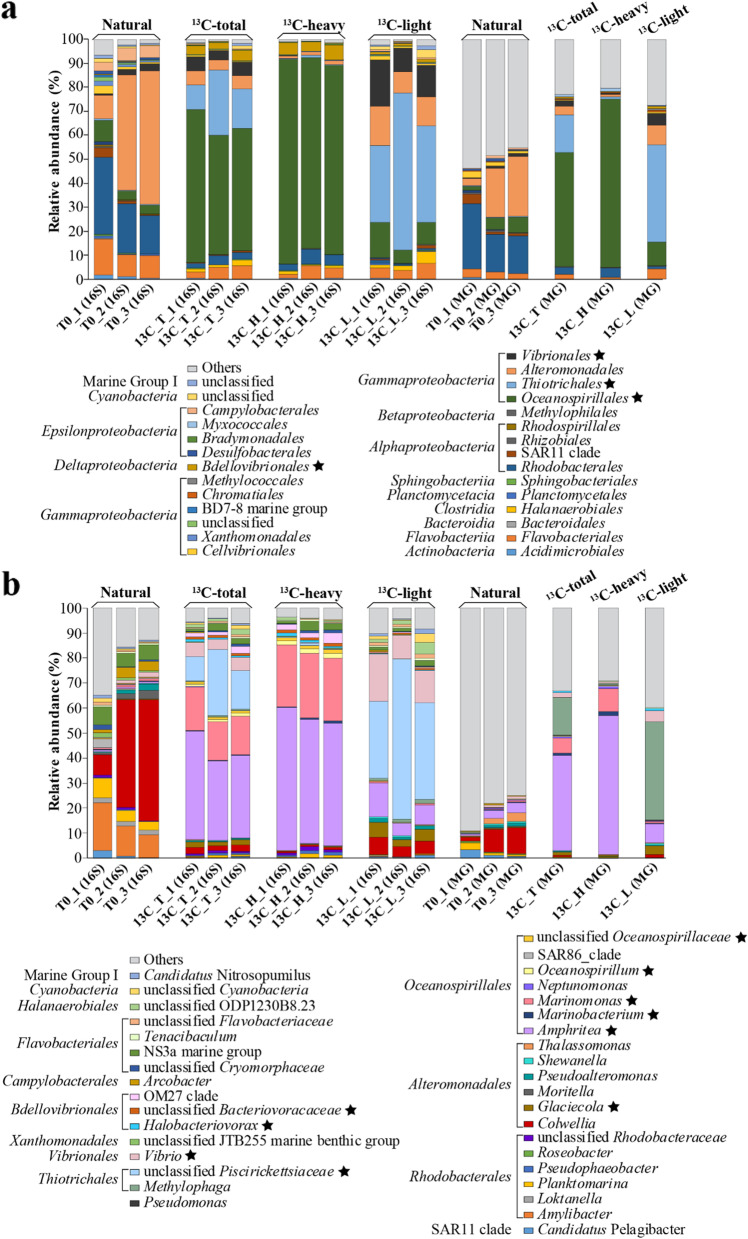
Fig. 3.
Microbial community profile of coastal seawater samples at order (a) and genus (b) levels. Bacterial diversity of the natural (T0) and labelled (heavy; H) and unlabelled (light; L) fractions of 13C-DMSP seawater incubations was analysed by 16S rRNA gene amplicon (16S) and metagenomics (MG) sequencing. “_1”, “_2” and “_3” after the sample name represent biological replicates. Biological replicates from 13C-heavy and 13C-light fractions were respectively combined before MG sequencing due to their highly similar 16S rRNA gene community profile shown by DGGE (Fig. S7). Only classes and genera with RA >0.5% in at least one of the conditions are represented. Statistically enriched genera in the incubations with 13C-labelled DMSP (13C_T) compared the natural (T0) samples based on 16S data are labelled with an asterisk. Classes and genera with RA <0.5% are grouped into “others”. For 16S and MG data of samples incubated with 12C-DMSP (controls), see Fig. S8