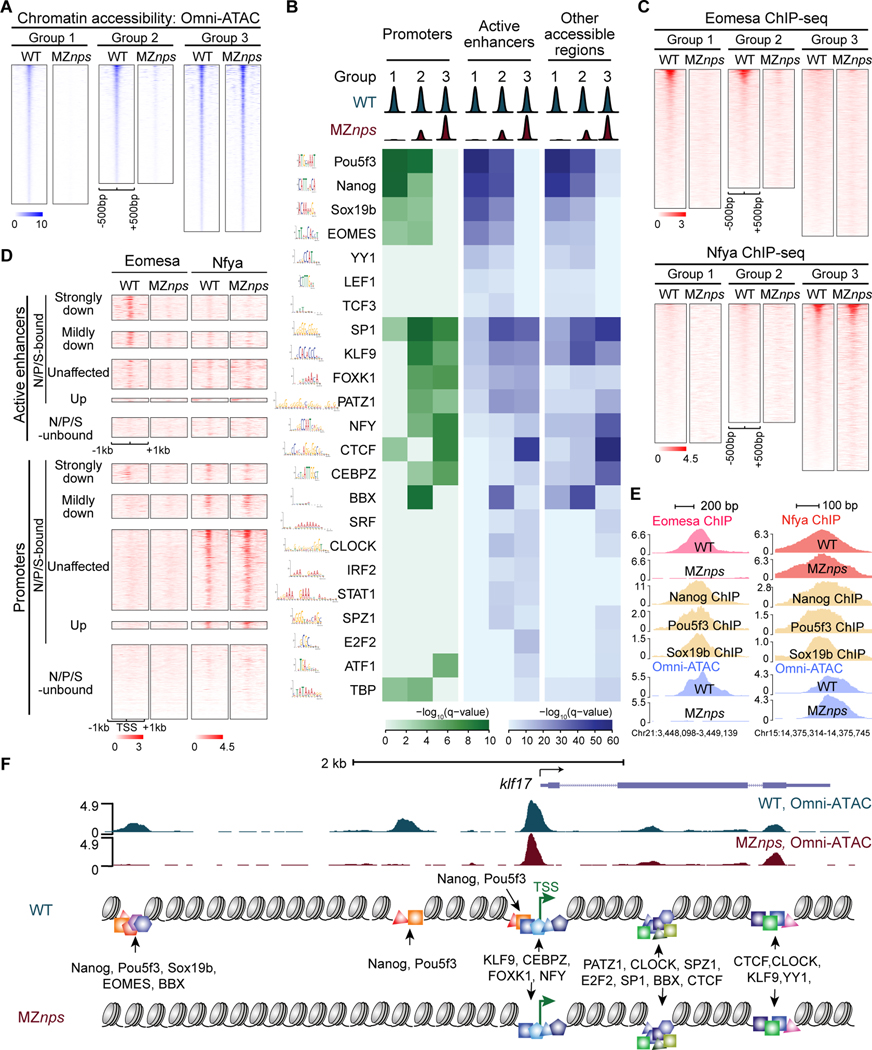
Figure 4. Identification of TF motifs enriched in accessible regions and their dependence on NPS.
(A) Heatmaps of Omni-ATAC signal at differentially affected accessible regions in wild-type (WT) and MZnps embryos. Heatmaps are centered at Omni-ATAC peak summits and ranked by the average Omni-ATAC signal intensity in WT. (B) Enrichment of representative TF motifs at promoters, active enhancers, and other accessible regions within the three groups of accessible regions from (A). (C) Heatmaps of Eomesa and Nfya ChIP-seq signal within the three groups of accessible regions from (A) in WT and MZnps embryos. Heatmaps are centered at the Omni-ATAC peak summits and ranked by average ChIP-seq signal in WT embryos. (D) Heatmaps showing Eomesa and Nfya levels at active enhancers and promoters of differentially affected zygotic genes. Heatmaps are centered at the TSS (promoters) and the summit of the Omni-ATAC peak (active enhancers) and ranked by average Pol II signal within 1kb of the TSS of the associated zygotic genes in WT embryos. (E) Representative genome tracks of TF binding and Omni-ATAC show that Eomesa binding depends on NPS pioneering activity, while Nfya binding is largely independent of NPS. (F) Diagram illustrating TF motifs located at the accessible regions near klf17. See also Figure S4.