TO THE EDITOR:
B-cell acute lymphocytic leukemia (B-ALL) is the most common childhood malignancy and is a rare leukemia in adults.1-4 B-ALL subtypes are distinguished by characteristic structural variants and mutations, which can correlate with responses to treatment.2-5 Cytogenetic and genomic analyses combined with expression profiling have identified the existence of up to 23 subtypes.4,6 Subtype assignment can extend and refine the current standards of risk stratification, and current standard of care incorporates some molecular classification to identify patients at higher risk.7,8 For instance, detection of BCR-ABL1 (Philadelphia (Ph) chromosome) indicates high-risk disease, and treatment can be modified to include an ABL1-targeting tyrosine kinase inhibitor such as imatinib,3 and ETV6-RUNX1 fusions can indicate a lower risk of relapse.7-9 Next-generation sequencing of RNA (RNA-seq) has been used to identify fusion genes, quantify gene expression, and perform variant calling to identify driver mutations.9,10 Although gene expression quantification is particularly useful for identifying molecular subtypes, there is currently no publicly available software for subtype classification with RNA-seq.
Here we present ALLSorts: a B-ALL gene expression classifier that attributes samples to 18 subtypes previously defined by Gu et al.4 ALLSorts has a novel hierarchical design that offers broader group classifications if more specific subtypes cannot be ascertained. Additionally, ALLSorts can attribute multiple subtypes to samples.11 When applied to both pediatric and adult cohorts, ALLSorts demonstrated high accuracy and was able to classify previously undefined samples. ALLSorts is open source and publicly available at https://github.com/Oshlack/ALLSorts.
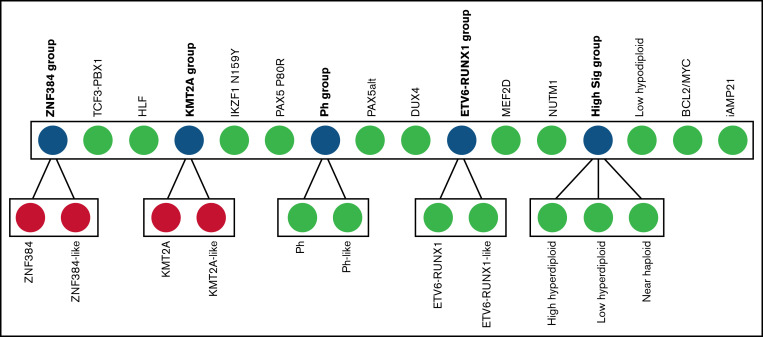
ALLSorts is a pretrained machine learning classifier that uses RNA-seq data to attribute B-ALL samples to 18 known subtypes. We developed ALLSorts by training a logistic regression classifier on a B-ALL dataset consisting of 1223 samples (supplemental Methods).4,6 ALLSorts uses an expression matrix for classification but can also accept FASTQ/FASTA or BAMs for conversion into this form. ALLSorts applies various processing steps to the data that are then input to a set of hierarchically organized logistic regression classifiers. Phenocopies are grouped into a meta-subtype with their mutational counterparts (Figure 1). These 5 meta-subtypes are as follows: ZNF384 group, KMT2A group, Ph group, ETV6-RUNX1 group, and high ploidy signature group (High Sig). The classifier first determines a sample's meta-subtype and then undertakes a more focused classification between the nested subtypes. The ZNF384-like and KMT2A-like subtypes contained too few training samples to confidently train a discriminator so default to their meta-subtype. This study was approved by the Royal Children's Hospital (RCH) Human Research Ethics Committee and the Peter Mac (PM) Human Research Ethics Committee and was performed in accordance with the Declaration of Helsinki.
Figure 1.
Overview of the ALLSorts classification architecture. In blue are the meta-subtypes that represent classes that have convergent or overlapping signals contained in their nested subtypes. Green nodes are terminal subtypes. Red nodes exist in the hierarchy, but classification currently terminates at the parent node because of a lack of training samples. CRLF2(non–Ph-like) is not included in this classification as its identification is better suited to alternate analysis. IGH-IL3 is also not included given only a single case was identified across all cohorts.
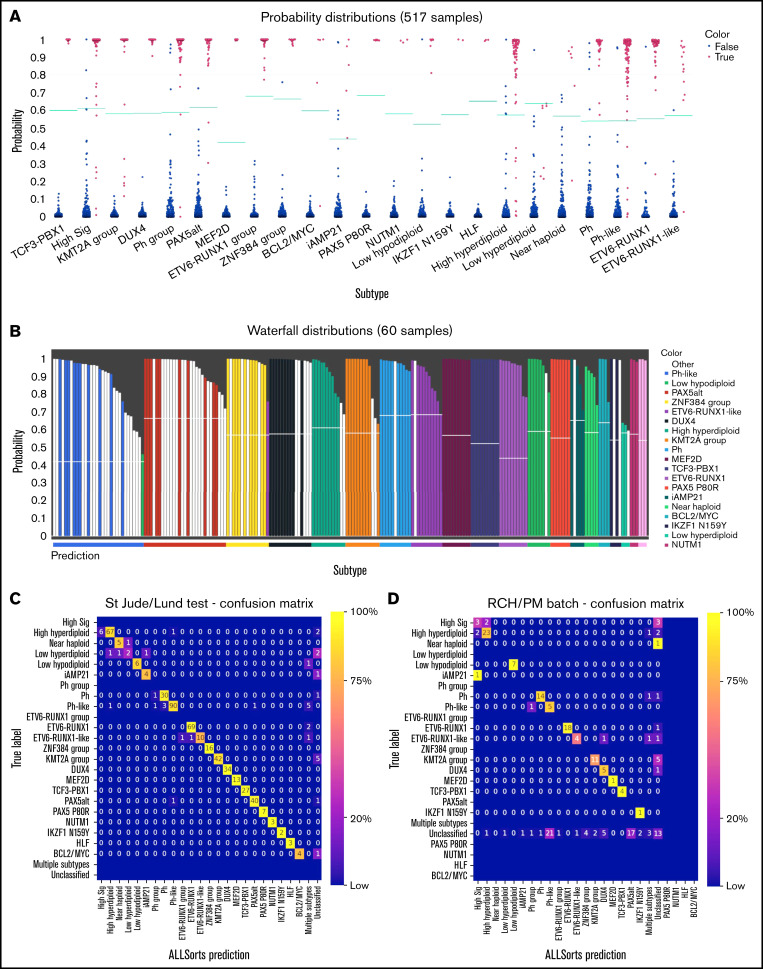
The outputs from ALLSorts for each sample are the subtypes with predicted probabilities. There are also 2 visualizations for validation and exploration of unclassified samples. The first visualization shows the sample’s probability of being a subtype relative to the predefined subtype threshold (Figure 2A). The second visualization, termed waterfall plots, compares the maximum subtype probability for each sample to the probabilities of samples known to belong to that subtype (Figure 2B).
Figure 2.
Validation of ALLsorts. (A) Predicted probabilities of St. Jude’s and Lund hold-out samples per subtype. For each sample, ALLSorts reports a probability for every subtype. Blue dots are samples negative for that subtype, and red are positive. The green lines are the subtype probability thresholds that were determined from the training data (supplemental Methods). (B) Waterfall plot of RCH and PM samples that were previously unclassified and assigned new subtypes by ALLSorts (white bars). The colored bars represent samples with positive classifications from the St. Jude’s/Lund held out set. The y axis shows the highest probability reported for the sample, and the x axis color is the prediction as made by ALLSorts. Samples with multiple subtypes are displayed in every subtype where a prediction is made. (C) Confusion matrix of the St. Jude’s/Lund held-out test data. A confusion matrix shows the performance of the classifier. The y axis represents the ground truth of each subtype, and the x -axis is the ALLSorts prediction. A perfect classification result would include no values off the diagonal. ALLSorts can predict samples to have multiple labels; these are reflected in the multiple subtypes category. Unclassified is where a sample’s probability did not exceed the threshold for any subtype. (D) Confusion matrix of the combined RCH and PM cohorts. The y axis represents the previous classification of each sample, and the x axis is the ALLSorts prediction. Rows without values indicate no subtype with that true label in the dataset.
The trained classifier was first applied to held-out test sets from the training cohorts (supplemental Table 5). ALLSorts was found to have an overall accuracy of 92% (Figure 2C). However, classification performance was unbalanced between subtypes. The best performance was for subtypes with a small number of clearly defined features, which were often partners in fusion genes. The highest levels of misclassification occurred for the subtypes with larger collections of features, especially the High Sig group. However, falling back to meta-subtypes in these cases, results in high accuracy. For example High Sig meta-subtype can be used with an accuracy of 93%. In addition, both Ph/Ph-like and ETV6-RUNX1/ETV6-RUNX1–like saw misclassifications to their phenotypic counterparts (Figure 2C). These observations highlight the utility of the novel hierarchical architecture in providing important classifications that can be explored and validated with complementary analysis or assays.
To validate ALLSorts on independent data, we applied it to 195 samples across 2 cohorts of pediatric and adult B-ALL from the RCH and PM, which displayed clear batch effects (supplemental Figure 5). These datasets have some previously defined subtype classifications from various combinations of fusion calling, karyotyping, genomic sequencing, or gene expression classification with an earlier machine learning approach.9
The initial accuracy of the classifier was 79%, assuming that all previous subtypes were correct but not including 74 (38%) previously unclassified samples. However, ALLsorts was able to newly classify 61 (82%) of these (Figure 2D). Forty-six of these new classifications were evaluated to be plausible using fusion calling,12-14 karyotyping, and genomic sequencing for variant calling. Ten samples were reclassified to a new subtype, of which 8 matched the previous meta-subtype label. There were 15 (7.7%) previously labeled samples, which ALLSorts assigned as unclassified. Six of these had tumor purities of less than 10%.
A full list of samples that had new classifications is provided, with any causative variants found (supplemental Table 7). Of these 86 samples, 63% had a plausible explanation that the ALLSorts classification was correct at least to the meta-subtype level, 8% were incorrect, 20% remained ambiguous in terms of evidence supporting or dismissing plausibility of the call, and 9% were defined as having low tumor purity (less than 10%). We found high accuracy of classification for tumor purities above 20% (supplemental Figures 6 and 7).
One unique feature of ALLSorts is its ability to classify samples into more than 1 subtype. The training cohorts included 117 samples that were previously described as having multiple subtypes based on both gene expression analysis and cytogenetics. Without specifically training ALLSorts to recognize samples exhibiting multiple subtypes, these samples were used to investigate the capacity for multilabel classification.
We found the probability of getting at least a single subtype correct is 86.31%, and 90.5% if including meta-subtypes. However, we only predicted both subtypes 26% of the time (supplemental Table 4). This implies that multiple label classification with ALLSorts can add further value of a classifier with little cost in performance. In the future, as further manual labeling of multilabel samples becomes available, these multilabel subtypes could be explicitly trained for.
In this study, we present ALLSorts, a B-ALL subtype classification tool that can precisely attribute samples to 18 subtypes and 5 meta-subtypes according to their RNA-seq measurements. This tool has been trained and validated with a combined cohort of more than 2300 samples and is offered for public use through Github. One novel contribution of ALLSorts is a hierarchical architecture representing subtypes and their phenocopies within a meta-subtype. Additionally, ALLSorts can also classify samples into more than one subtype.
A key component of this study was testing the predictions of the software across validation cohorts to verify the robustness of the classifier. We found that the overall accuracy in the combined independent cohort was between 84% and 92% (supplemental Table 3). ALLSorts has the ability to retrain the classifier as more samples become available, which will allow classification of subtypes that currently have relatively low numbers of samples, such as BCL2/MYC. Although gene counts are clearly useful in determining the subtype, a more refined method that uses nuanced aspects of the data such as transcript quantification could provide increased performance. Complementary analysis methods such as fusion detection should be used in conjunction with ALLSorts for a broader picture. However, we clearly demonstrate that ALLSorts is capable of high classification accuracy across an extensive set of subtypes.
In summary, ALLSorts is an accurate, comprehensive, and freely available classification tool for determining subtypes of B-ALL.
Supplementary Material
The full-text version of this article contains a data supplement.
Acknowledgments
Acknowledgments: Tumor samples and coded data were supplied by the Children’s Cancer Centre Tissue Bank at the Murdoch Children’s Research Institute and The Royal Children’s Hospital (www.mcri.edu.au/childrenscancercentretissuebank). Establishment and running of the Children’s Cancer Centre Tissue Bank is made possible through generous support by Cancer In Kids @ RCH, The Royal Children’s Hospital Foundation, and the Murdoch Children’s Research Institute. Raw gene expression counts for B-ALL tumor samples used for analysis in this study were obtained from St. Jude Cloud (https://www.stjude.cloud), which is a publicly accessible pediatric genomic data resource requiring approval for controlled data access.
The authors acknowledge the support of the SCOR Grant (7015-18) from the Lymphoma and Leukemia Society and of Perpetual Trustees and the Samuel Nissen Foundation.
This work was supported by grants from the Wilson Centre for Lymphoma Genomics and the Snowdome Foundation. This work was funded by National Health and Medical Research Council project grant GNT1140626.
Contribution: B.S. conceptualized the study, performed the formal analysis, created the methodology, provided software, visualized the study, wrote the original draft, and reviewed and edited the manuscript; A.O. conceptualized the study, supervised the study, created the methodology, visualized the study, wrote the original draft, and reviewed and edited the manuscript; N.M.D. conceptualized the study, supervised the study, created the methodology, wrote the original draft, and reviewed and edited the manuscript; L.M.B. provided clinical expertise and reviewed and edited the manuscript; G.L.R. provided sample and clinical expertise and reviewed and edited the manuscript; A.L. provided bioinformatics support and reviewed and edited the manuscript; H.J.K. provided biological expertise and reviewed and edited the manuscript; L.E.L. provided orthogonal clinical information and reviewed and edited the manuscript; I.J.M. provided biological expertise and reviewed and edited the manuscript; P.B. provided clinical expertise and reviewed and edited the manuscript; and P.G.E. provided clinical expertise and reviewed and edited the manuscript.
Conflict-of-interest disclosure: The authors declare no competing financial interests.
Correspondence: Alicia Oshlack, Computational Biology Program, Peter MacCallum Cancer Centre, Parkville, VIC 3000, Australia; e-mail: alicia.oshlack@petermac.org.
References
- 1.Hunger SP, Mullighan CG. Redefining ALL classification: toward detecting high-risk ALL and implementing precision medicine. Blood. 2015;125(26):3977-3987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Terwilliger T, Abdul-Hay M. Acute lymphoblastic leukemia: a comprehensive review and 2017 update. Blood Cancer J. 2017; 7(6):e577. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Inaba H, Greaves M, Mullighan CG. Acute lymphoblastic leukaemia. Lancet. 2013;381(9881):1943-1955. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Gu Z, Churchman ML, Roberts KG, et al. PAX5-driven subtypes of B-progenitor acute lymphoblastic leukemia. Nat Genet. 2019;51(2):296-307. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Paietta E, Roberts KG, Wang V, et al. Molecular classification improves risk assessment in adult BCR-ABL1-negative B-ALL. Blood. 2021;138(11):948-958. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Lilljebjörn H, Henningsson R, Hyrenius-Wittsten A, et al. Identification of ETV6-RUNX1-like and DUX4-rearranged subtypes in paediatric B-cell precursor acute lymphoblastic leukaemia. Nat Commun. 2016;7:11790. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Schultz KR, Pullen DJ, Sather HN, et al. Risk- and response-based classification of childhood B-precursor acute lymphoblastic leukemia: a combined analysis of prognostic markers from the Pediatric Oncology Group (POG) and Children’s Cancer Group (CCG). Blood. 2007;109(3):926-935. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Inaba H, Azzato EM, Mullighan CG; Jude Children’s Research Hospital Approach . Integration of next-generation sequencing to treat acute lymphoblastic leukemia with targetable lesions: the St. Jude Children’s Research Hospital Approach. Front Pediatr. 2017;5:258. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Brown LM, Lonsdale A, Zhu A, et al. The application of RNA sequencing for the diagnosis and genomic classification of pediatric acute lymphoblastic leukemia. Blood Adv. 2020;4(5): 930-942. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Byron SA, Van Keuren-Jensen KR, Engelthaler DM, Carpten JD, Craig DW. Translating RNA sequencing into clinical diagnostics: opportunities and challenges. Nat Rev Genet. 2016;17(5):257-271. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Nordlund J, Bäcklin CL, Zachariadis V, et al. DNA methylation-based subtype prediction for pediatric acute lymphoblastic leukemia. Clin Epigenetics. 2015;7:11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Nicorici D, Şatalan M, Edgren H, et al. FusionCatcher: a tool for finding somatic fusion genes in paired-end RNA-sequencing data. bioRxiv. 2014. doi: 10.1101/011650. [DOI]
- 13.Davidson NM, Majewski IJ, Oshlack A. JAFFA: high sensitivity transcriptome-focused fusion gene detection. Genome Med. 2015;7(1):43. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Uhrig S, Ellermann J, Walther T, et al. Accurate and efficient detection of gene fusions from RNA sequencing data. Genome Res. 2021;31(3):448-460. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.