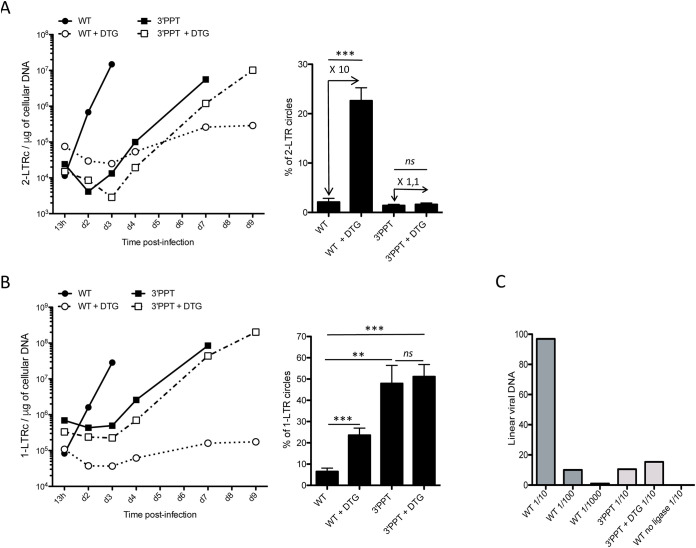
FIG 2.
Characterization of unintegrated viral DNA (linear and 2-LTR and 1-LTR circles) genomes following infection with the WT or 3′-PPT mutant virus. Samples from the previously depicted experiment (Fig. 1) using MT4 T-cells infected with the WT or 3′-PPT mutant virus, in the presence or absence of 1 μM DTG, were analyzed by qPCR to quantify circular genomes: (A) Left panel: time course of 2-LTR circle formation. Right panel: average percentage of 2-LTR circles for all time points analyzed. The fold of 2-LTR circle accumulation due to DTG treatment is indicated in the Figure. (B) Left panel: time course of 1-LTR circle formation. Right panel: average percentage of 1-LTR circles for all time points analyzed. (C) Quantification of linear viral DNA at 13h postinfection by ligation-mediated PCR. Several dilutions have been tested for each sample (1/10, 1/100, 1/1000 for the WT and 1/10 for the 3′-PPT and 3′-PPT + DTG). In addition, as a negative control, a sample of WT was processed without the ligase. All results were obtained from three independent experiments (mean ± SEM). Statistical differences were calculated using Student’s test (*, P < 0.05; **, P < 0.005; ***, P < 0.001; ns P > 0.05).