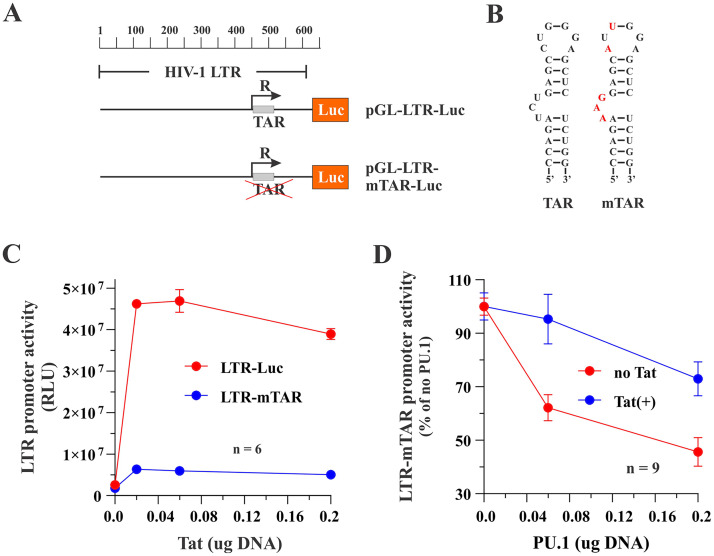
FIG 9.
Tat binding to the TAR stem-loop structure of the HIV-1 LTR is not required for PU.1-mediated inhibition of the HIV-1 LTR promoter activity. (A) Schematic representation of the constructs used in this experiment. (B) To minimize binding of Tat to the LTR, the TAR stem-loop structure in pGL-LTR-mTAR-Luc was mutated in the bulge-and-loop region, as previously reported, (52) using site-directed mutagenesis. Mutated residues are shown in red. (C) HEK293T cells were transfected in 24-well plates with pGL-LTR-Luc (0.1 μg) or pGL-LTR-mTAR-Luc (0.1 μg) together with increasing amounts of pTat101 (0 μg, 0.02 μg, 0.06 μg, and 0.2 μg). Production of luciferase was assessed 2 days later. Means and error bars reflecting SEM calculated from six independent transfections are shown. LTR promoter activity was statistically significantly different between 0 μg and 0.2 μg Tat for both LTR-Luc (P = 0.001) and LTR-mTAR (P = 0.009). (D) HEK293T cells were transfected in 24-well plates with pGL-LTR-mTAR-Luc (0.1 μg) in the absence (red circles) or presence (blue circles) of 0.1 μg pTat101 and increasing amounts of PU.1 DNA (0 μg, 0.06 μg, and 0.2 μg). Production of luciferase was assessed 2 days later. Means and error bars reflecting SEM calculated from nine independent transfections are shown. LTR-mTAR-driven luciferase activity by PU.1 was statistically significantly different between 0 μg and 0.2 μg PU.1 both in the presence (P = 0.006) and absence (P < 0.001) of Tat. The slope of the inhibition curves was significant in the presence or absence of Tat, although the curves were not statistically significantly different from each other. For details on the statistical analyses, see Fig. S1 in the supplemental material.