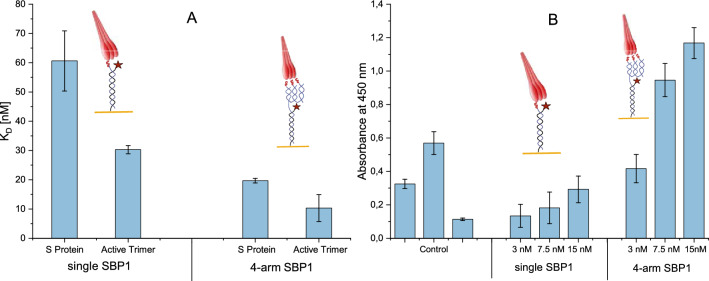
Figure 3.
Spike protein binding to three or one peptide per DNA nanostructure. (A) Results from the switchSENSE measurements of the dissociation constants KD for the binding interaction of “SARS-CoV-2 S protein” and “SARS-CoV-2 S protein, active trimer” by Acro Biosystems to the monovalent peptide presenting DNA (single SBP1) and the trimeric peptide presenting DNA structure (4-arm SBP1). Trimeric peptide presentation reduces the dissociation constant by a third compared to the monovalent peptide presentation. The “SARS-CoV-2 S protein, active trimer” binds more strongly than the “SARS-CoV-2 S protein”, presumably due to variations in glycosylation and amino acid sequence variations. (B) Results from ELISA for a control, immobilized DNA with one peptide or three peptides and subsequent binding analysis of the “SARS-CoV-2 S protein, active trimer”. Here as well the trimeric presentation results in a significant increase of the signal compared to the monovalent peptide presentation. Although the experiment was repeated multiple times, the results shown here come from one well plate to allow for comparability.