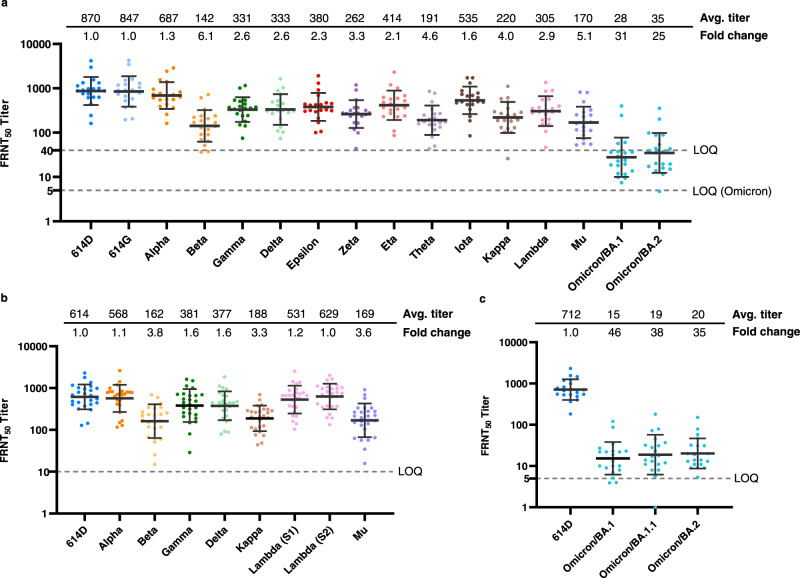
Fig. 2. Neutralizing activity of mRNA vaccinee sera (post-second dose) against live SARS-CoV-2 viruses.
Each dot represents the neutralizing titer (FRNT50) of an individual serum sample against a specific SARS-CoV-2 virus, which is labeled on the X-axis. Different colors represent the progenitor virus or different variants (specific mutations provided in Supplementary Table 1). Vaccinee serum samples were obtained from people who were vaccinated 2–6 weeks post second dose. The geometric mean FRNT50 titers are represented in the graph as bars on top of the dots with geometric standard deviation. The geometric mean FRNT50 titers (avg. titer) and average fold changes relative to reference virus 614D (set as 1-fold) are shown on the top of the graph. For each variant, the average fold change is the geometric mean of the individual FRNT50 ratios (614D/variant) calculated for each serum sample. Dashed line represents the limit of quantitation (LOQ). For statistical analysis, a two-tailed Wilcoxon matched-pairs signed-rank test was performed by comparing each variant with 614D. Test statistics and P value are summarized in Supplementary Table 2. Source data are provided as a Source Data file. a Representative reporter viruses of all past and current WHO designated SARS-CoV-2 VOCs and VOIs were tested. N = 20 biologically independent sera examined over 16 viruses. LOQ = 5 for Omicron and LOQ = 40 for all other viruses. The average fold changes of all viruses differ significantly (P < 0.0001) from 614D, except for 614G (P = 0.8124). b VOCs and selected VOIs isolated from clinical specimens were tested. N = 26 biologically independent sera examined over 9 viruses. The average fold changes of all viruses differ significantly (P < 0.0001) from 614D, except for Alpha (P = 0.4833) and the two Lambda viruses (Lambda S1 and S2, P = 0.0796 and 0.7265, respectively). c Omicron subvariant BA.1, BA.1.1, and BA.2 isolated from clinical specimens were tested. N = 20 biologically independent sera examined over four viruses. The average fold changes of all viruses differ significantly (P < 0.0001) from 614D.