FIGURE 2.
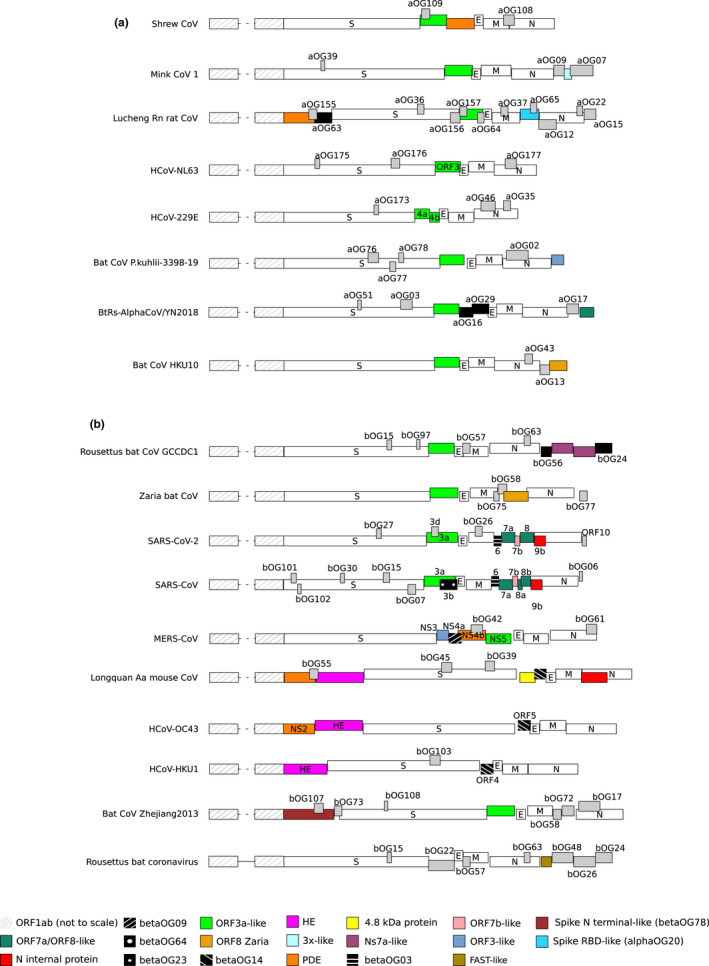
AlphaCoV and betaCoV genome organization. A schematic genome organization of representative alphaCoV (a) and betaCoV (b) genomes. Super‐groups and relevant orthogroups are coloured as shown in the key; known and unknown orthogroups are coloured in black and grey, respectively, and orthogroup names are reported. For all viruses, ORF1ab is not shown to scale and structural proteins are coloured in white. For human coronaviruses, accessory protein names are also reported. Viruses reported in the figure are as follows: Common shrew coronavirus Tibet‐2014: KY370053; mink coronavirus 1: MN535737; Lucheng Rn rat coronavirus: MT820627; HCoV‐NL63: NC_005831; HCoV‐229E: NC_002645; Alphacoronavirus bat‐CoV/P.kuhlii/Italy/3398–19/2015: NC_046964; coronavirus BtRs‐AlphaCoV/YN2018: MK211373; Rousettus bat coronavirus HKU10: NC_018871; Rousettus bat coronavirus GCCDC1: MT350598; Zaria bat coronavirus: HQ166910; SARS‐CoV‐2: NC_045512; SARS‐CoV: NC_004718; MERS‐CoV: NC_019843; Longquan aa mouse coronavirus: KF294357; HCoV‐OC43: NC_006213; HCoV‐HKU1: NC_006577; bat Hp‐betacoronavirus/Zhejiang2013: NC_025217; Rousettus bat coronavirus isolate GCCDC1 356: NC_030886
