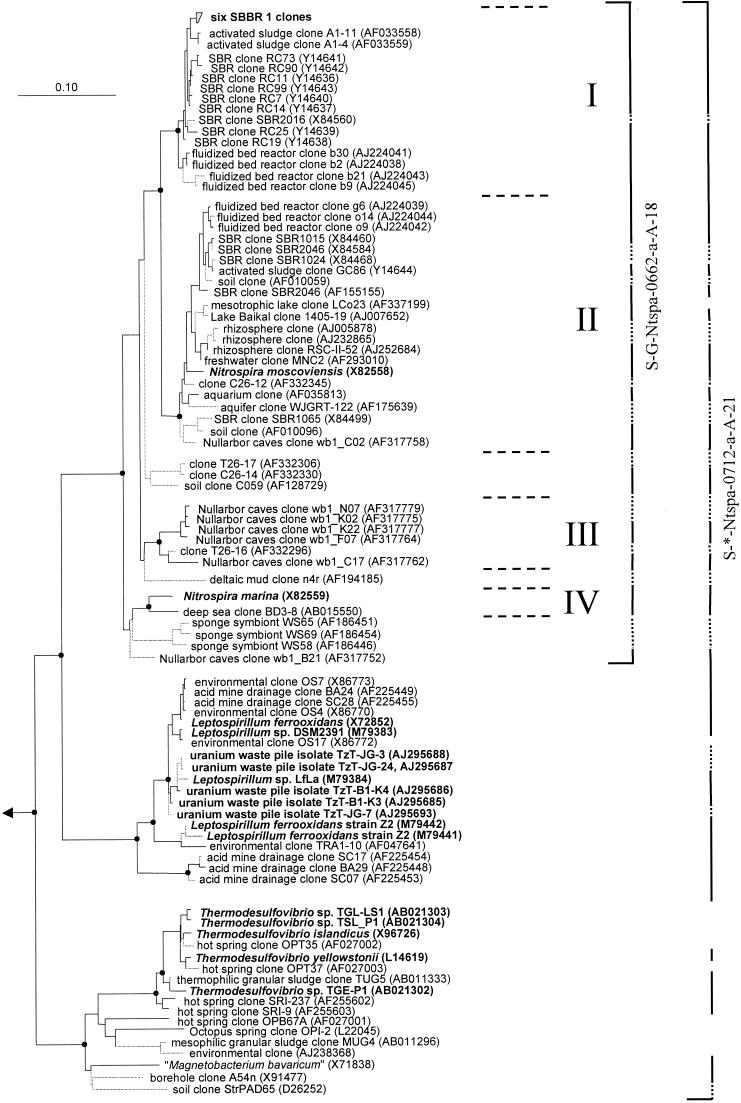
FIG. 1.
Phylogenetic tree of the phylum Nitrospira based on comparative analysis of 16S rRNA sequences. The basic tree topology was determined by maximum-likelihood analysis of all sequences longer than 1,300 nucleotides. Shorter sequences were successively added by use of the ARB_PARSIMONY module of the ARB program without changing the overall tree topology. Branches leading to sequences shorter than 1,000 nucleotides are dotted to point out that the exact affiliation of these sequences cannot be determined. Black spots on tree nodes symbolize high-parsimony bootstrap support above 90% based on 100 iterations. The scale bar indicates 0.1 estimated change per nucleotide. Sequences of Nitrospira-like bacteria retrieved in this study from reactor SBBR 1 and sequences that belong to isolated strains are in boldface. The four sublineages of the genus Nitrospira are delimited by horizontal dashed lines and numbered I to IV. The brackets illustrate the coverage of the 16S rRNA-targeted oligonucleotide probes developed in this study. Dotted bracket segments indicate that the corresponding partial sequences do not include the probe target site. Brackets are interrupted where sequences are not targeted by the respective probe.