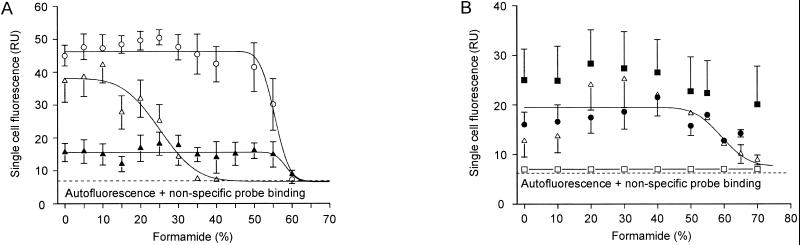
FIG. 3.
Probe dissociation profiles of the oligonucleotide probes developed in this study with reference organisms under increasingly stringent hybridization and washing conditions. For each data point, the mean fluorescence intensity of at least 100 cells was determined. Regression curves were calculated by the plotting software based on a sigmoidal curve fit model. Error bars indicate 1 standard deviation. Error bars that are smaller than the marker symbols are not shown. (A) Hybridization of target organism N. moscoviensis with probe S-G-Ntspa-0662-a-A-18 in the presence of Comp-Ntspa-0662 (▴). Hybridization of nontarget bacterium B. stearothermophilus with probe S-G-Ntspa-0662-a-A-18 without (○) and with (Δ) competitor Comp-Ntspa-0662. (B) Hybridization of target organisms N. moscoviensis (●) and L. ferrooxidans (■) with probe S-*-Ntspa-0712-a-A-21 in the presence of competitor Comp-Ntspa-0712. Hybridization of nontarget bacterium D. desulfuricans with probe S-*-Ntspa-0712-a-A-21 without (▵) and with (□) competitor Comp-Ntspa-0712. The regression curve refers to the data points obtained for D. desulfuricans without addition of the competitor. RU, relative units.