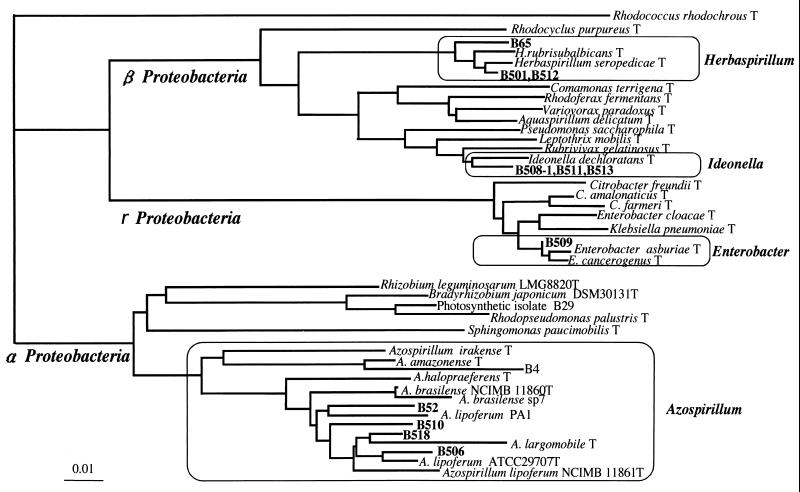
FIG. 1.
Phylogenetic tree showing relatedness of nitrogen-fixing bacteria isolated from rice and their relatives in the Proteobacteria by neighbor-joining grouping of the aligned sequences of the 16S rRNA gene. Strains B4 and B29 are nitrogen-fixing bacteria previously isolated using nutrient agar medium from surface-sterilized culms of wild rice (9). To construct the phylogenetic tree, we used the 16S rRNA sequences in the DDBJ/EMBL/GenBank database from the following bacteria (accession numbers given in parentheses): Rhodococcus rhodochrous (X79288), Rhodocyclus purpureus (M34132), Herbaspirillum rubrisubalbicans (AB021424), Herbaspirillum seropedicae (Y10146), Comamonas terrigena (AB021418), Rhodoferax fermentans (D16211), Variovorax paradoxus (D30793), Aquaspirillum delicatun (AF078756), Pseudomonas saccharophila (AB021407), Leptothrix mobilis (X97071), Rubrivivax gelatinosus (D16213), Ideonella dechloratans (X72724), Citrobacter freundii (M59291), Citrobacter amalonaticus (AF025370), Citrobacter farmeri (AF025371), Enterobacter cloacae (AJ251469), Klebsiella pneumoniae (AF130981), Enterobacter asburiae (AB004744), Enterobacter cancerogenus (Z96078), Rhizobium leguminosarum (X67227), Bradyrhizobium japonicum (X87272), Rhodopseudomonas palustris (D25312), Sphingomonas paucimobilis (U37337), Azospirillum irakense (Z29583), Azospirillum amazonense (X79735), Azospirillum halopraeferens (X79731), Azospirillum brasilense NCIMB 11860T (Z29617), Azospirillum brasilense sp7 (X79739), Azospirillum lipoferum PA1 (X79738), Azospirillum largomobile (X90759), Azospirillum lipoferum ATCC 29707T (M59061), and Azospirillum lipoferum NCIMB11861T (Z29619). Bar, 0.01 base substitution per nucleotide.