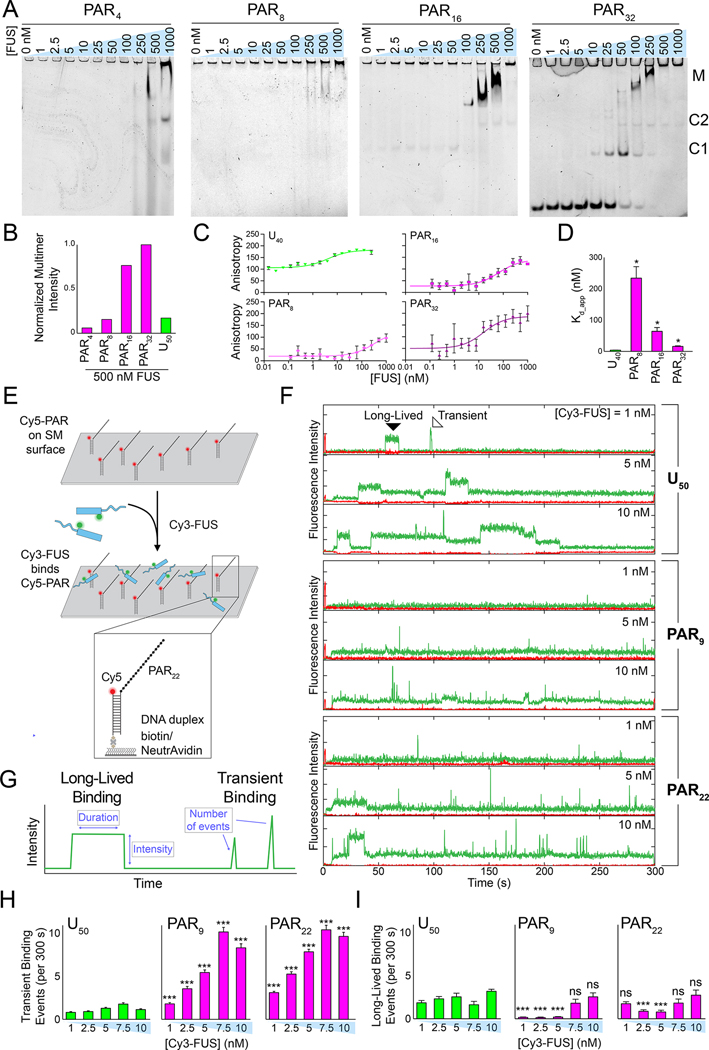
Figure 2: PAR interacts with FUS primarily through transient interactions.
(A) Electrophoretic mobility shift assays (EMSAs) of increasing concentrations of FUS incubated with different lengths of 1 nM PAR. The annotations denote the following complexes: M = multimers, C2 = complex 2 (FUS dimer), C1 = complex 1 (FUS monomer). (B) Quantification of normalized aggregation observed on an EMSA gel of all PARs and U50 RNA incubated with FUS (see Figure S2A). (C) Fluorescence anisotropy binding isotherms of increasing concentrations of FUS incubated with 10 nM Cy5-PAR or Cy5-U40. Error bars represent standard error of the mean (n = 3), and best fit lines were plotted by fitting with a four-parameter total binding equation in Prism 8. (D) Apparent FUS–PAR/RNA Kd values derived from the fits in (C). Error is standard error of the mean (n = 3), and each PAR Kd was pairwise compared with the Kd of U40 using Welch’s t-test where * = p < 0.05. (E) Schematic of the single-molecule nucleation assay with pdPAR constructs. (F) Representative single-molecule traces of increasing concentrations of Cy3-FUS added to pdRNA or pdPAR constructs. Examples of long-lived and transient binding interactions are denoted in the first trace. (G) Schematic of analysis of single-molecule traces. (H) Quantification of the average number of transient binding events per trace (i.e. per 300 s) for FUS with the pdPAR and pdRNA constructs. Error bars are standard error of the mean (n > 100), and significance of pairwise pdU50-pdPAR averages was calculated using Welch’s t-test. (I) Same as H, but with long-lived binding events.