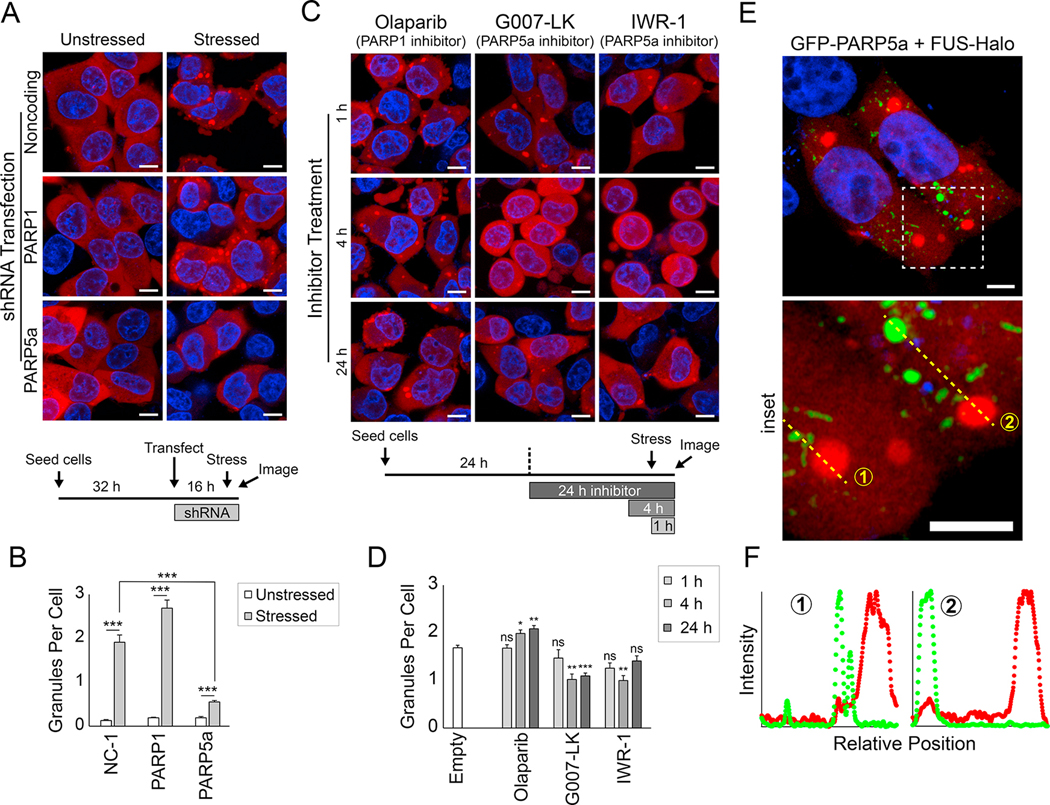
Figure 4: PARP5a activity promotes FUS condensation in cells.
(A) Representative confocal images of FUS-Halo SH-SY5Y cells treated with shRNAs for the indicated genes (see STAR Methods and timeline at the bottom of the panel). FUS was visualized with JF549 (red), and nuclei were visualized with Hoechst (blue). Cells were stressed with sodium arsenite for 1 h, and cells were live-imaged at 37 °C. The scale bar is 8 μm. (B) Quantification of the FUS granules per cell in (A). The error bars denote standard error of the mean (n > 30), and Welch’s t-test was used to calculate the significance of the indicated pairwise comparisons. NC-1, noncoding shRNA control. (C) Same as (A) but instead of transfected shRNAs, cells were treated with PARP inhibitors for the indicated time period (see timeline at the bottom of the panel). All images show live cells that were stressed with sodium arsenite for 1 h. (D) Same as (B) but for the data in (C). Significance was calculated for each condition compared to untreated FUS-Halo cells. (E) Airyscan confocal images of FUS-Halo cells transfected with GFP-PARP5a (green). The inset shows the dashed-line box. The scale bar is 5 μm. (F) Normalized intensity plots show the enrichment of the FUS-Halo (red) and GFP-PARP5a channels for the yellow dashed lines in the inset in (E).