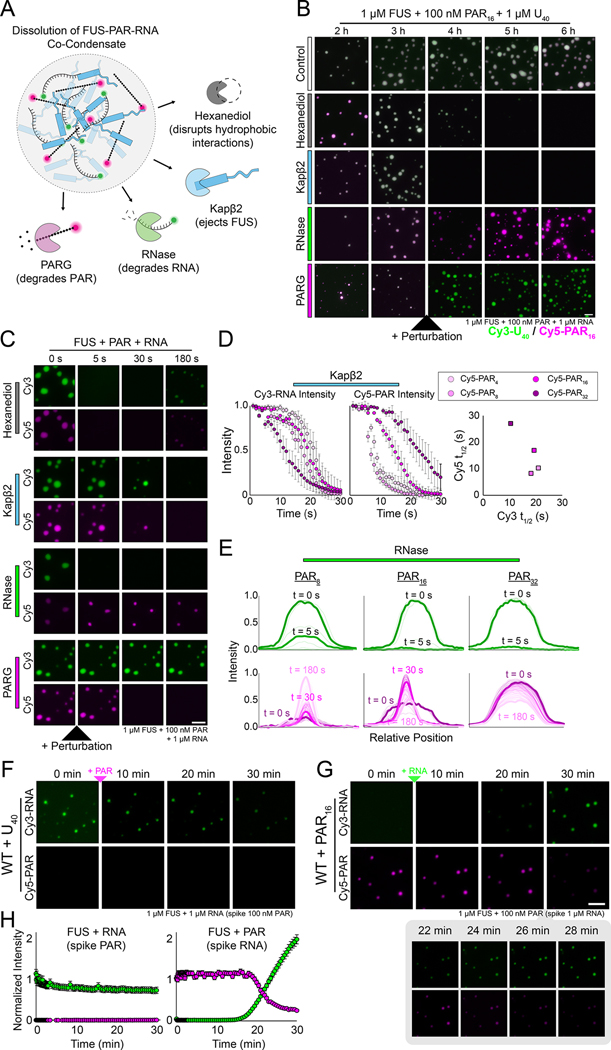
Figure 7: The architectural network of FUS–RNA–PAR co-condensates.
(A) Schematic showing how FUS–RNA–PAR co-condensates were treated with various proteins/chemicals. (B) Representative fluorescence wide-field images of FUS–RNA–PAR co-condensates before and after treatment. The Cy5 (magenta) signal is PAR and the Cy3 signal (green) is RNA. The scale bar is 5 μm. (C) Stills from representative dissolution videos of FUS–RNA–PAR16 co-condensates. The Cy5 (magenta) signal is PAR and the Cy3 signal (green) is RNA. The treatments were added just after the 0 s time point. For Kapβ2, time points are shown for 0, 5, 15, and 30 s. The scale bar is 5 μm. (D) Intensity plots of the Cy3-RNA and Cy5-PAR signals for each of the indicated co-condensate reactions treated with Kapβ2. Error bars denote standard deviation (n=10). The plot on the right shows the fitted t1/2 value for the Cy3 and Cy5 dissolution intensity curves. (E) Representative intensity plots of individual FUS–RNA–PAR condensates treated with RNase. The lighter lines indicate later time points during the video acquisition, and bolded lines are highlighted times as indicated. (F) Wide-field fluorescence images of FUS-Cy3-RNA droplets immediately after adding Cy5-PAR. (G) Same as (F) but for FUS-Cy5-PAR droplets after adding Cy3-RNA. The inset below shows more timepoints of the Cy3-RNA displacement of Cy5-PAR. The scale bar is 5 μm, which also applies to panel (F). (H) Quantification of the Cy3 and Cy5 fluorescence signals in (F) and (G).