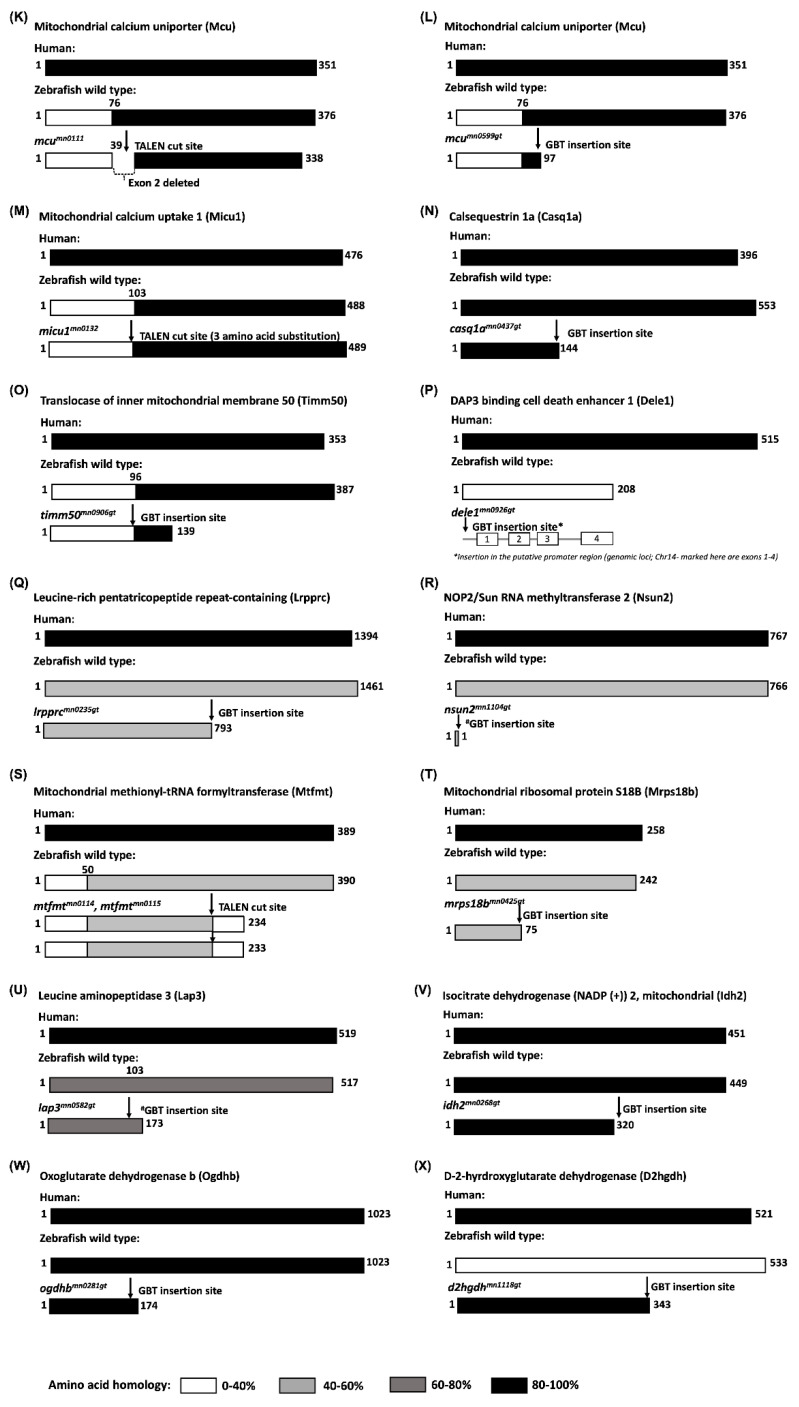
Figure 3.
A depiction of homology of the mutants created for proteins involved in different mitochondrial resident pathways: (A,B): Complex 1 of mitochondrial respiratory chain; (A): NADH: ubiquinone oxidoreductase complex assembly factor 6 (Ndufaf6); (B): NADH: ubiquinone oxidoreductase subunit S4 (Ndufs4); (C): Complex 2 of mitochondrial respiratory chain; (C): Succinate dehydrogenase complex flavoprotein subunit A (Sdha); D-E: Biosynthesis of coenzyme Q; (D): Coenzyme Q2, polyprenyltransferase (Coq2) (E): Decaprenyl-diphosphate synthase subunit 2 (Pdss2); (F): Complex 3 of mitochondrial respiratory chain; (F): Ubiquinol–cytochrome c reductase complex III subunit VIII (Uqcrq); (G,H): Complex 4 of mitochondrial respiratory chain; (G): SURF1 cytochrome C oxidase assembly factor (Surf1); (H): Cytochrome C oxidase assembly factor heme A:farnesyltransferase COX10 (Cox10); (I,J): Complex 5 of mitochondrial respiratory chain; (I): Transmembrane protein 70 (Tmem70); (J): ATP synthase F1 subunit epsilon (Atp5f1e); (K–N): Mitochondrial calcium homeostasis; (K): Mitochondrial calcium uniporter (Mcu) generated by TALEN and (L); by GBT; (M): Mitochondrial calcium uptake 1 (Micu1); (N): Calsequestrin 1 (Casq1a); (O): Mitochondrial protein import; (O): Translocase of inner mitochondrial membrane 50 (Timm50); (P): Stress response; (P): DAP3 binding cell death enhancer 1 (Dele1) (GBT insertion site is in the putative promoter region, site is depicted upstream of exons of dele1 gene in chromosome 14; (Q–U): Mitochondrial genome maintenance; (Q): Leucine-rich pentatricopeptide repeat containing (Lrpprc); (R): NOP2/Sun RNA methyltransferase 2 (Nsun2) #candidate line; (S): Mitochondrial methionyl-tRNA formyltransferase (Mtfmt); (T): Mitochondrial ribosomal protein S18B (Mrps18b); (U): Leucine aminopeptidase 3 (Lap3) #candidate line; (V–X): Mitochondrial metabolite synthesis; (V): Isocitrate dehydrogenase (NADP(+)) 2, mitochondrial (Idh2); (W): Oxoglutarate dehydrogenase b (Ogdhb); (X): D-2-hydroxyglutarate dehydrogenase (D2hgdh).