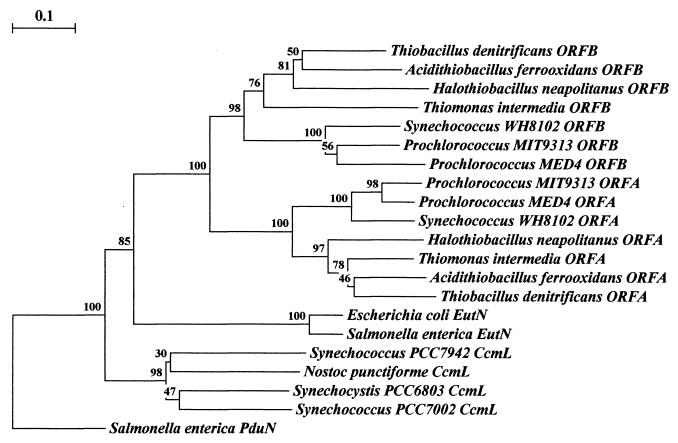
FIG. 4.
Phylogenetic tree of the ORFA and ORFB polypeptides and their relatives. Rooting was done arbitrarily through Salmonella enterica serovar Typhimurium PduN, which groups individually and away from the other families of polypeptides. Clustering the polypeptides into groupings respective of species physiology may be seen among the cyanobacteria, the thiobacilli, and the enteric polypeptides. Uniform clustering of protein homologs by relative physiology occurs with a high degree of confidence. Of the major branches of protein groupings, all are represented by bootstrap values greater than 75%. The data for each bootstrap value were resampled 1,000 times, with the evolutionary distance scale in the upper left-hand corner representing the number of fixed substitutions per site, as a measure of horizontal distance. Sequence alignment was performed with ClustalW 1.8 (70) and phylogenetic trees were constructed with Treecon (72, 73).