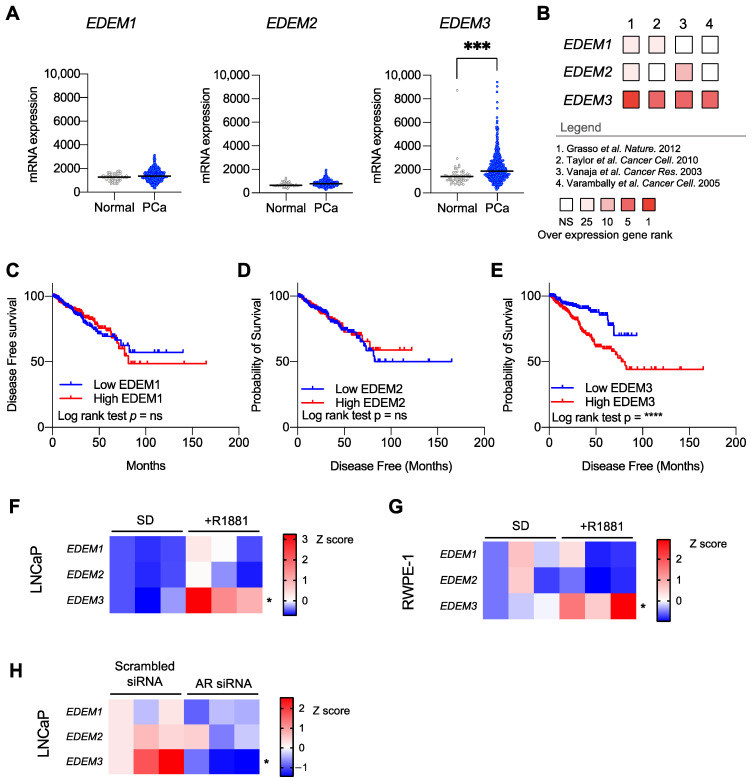
Figure 1.
EDEM3 upregulation is associated with a reduced disease-free survival in prostate cancer (PCa). (A) Analysis of EDEM gene expression in The Cancer Genome Atlas (TCGA) prostate adenocarcinoma cohort (n = 497) compared with normal prostate samples (52). (B) Meta-analysis of EDEM1/2/3 gene expression in prostate cancer. Four independent prostate cancer gene expression data sets (n = 366) were analysed for EDEM1/2/3 gene expression in prostate cancer patients compared with patients with normal prostates. Data shown are of overexpression gene rank in each dataset. Legend includes details of the independent cohorts. NS = not significant, 25 = top 25%, 10 = top 10%, 5 = top 5% and 1 = top 1%. Data accessed using Oncomine [29,30,31,32]. (C–E) Kaplan–Meier plot showing disease-free survival for prostate cancer patients stratified based on low (bottom 50%) or high (top 50%) EDEM3 expression. Analysis includes 471 prostate cancer patients from TCGA PRAD cohort, accessed via CBioPortal. p value was calculated by log rank test. (F,G) Heatmaps showing mRNA levels of EDEM1/2/3 in both LNCaP and RWPE-1 cultured in either steroid-depleted (SD) conditions or stimulated with 10 nM R1881 for 24 h. n = 3 and data are presented as z-scores. (H) Heatmap showing mRNA levels of EDEM1/2/3 in LNCaP cells following androgen receptor siRNA knockdown, compared with scrambled siRNA control. n = 3 and data are shown as z-scores. p values were calculated using a two-tailed unpaired t-test. * p < 0.05, *** p < 0.001, **** p < 0.0001.