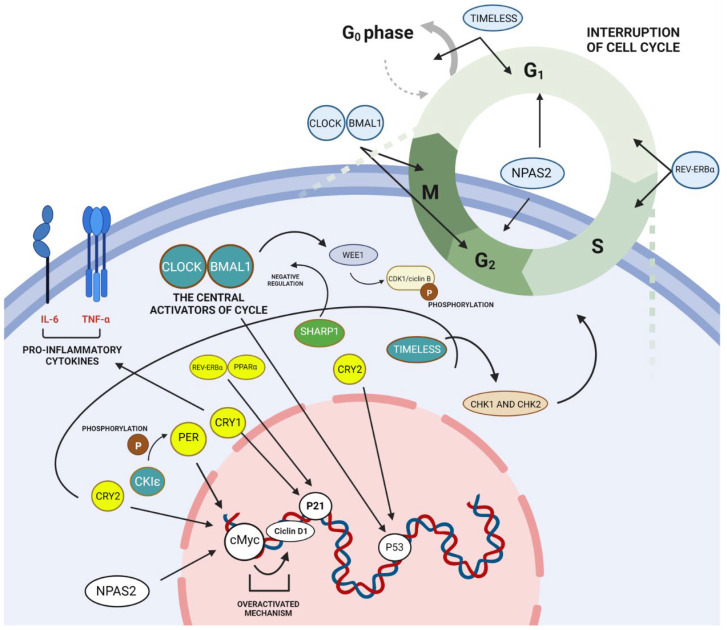
Figure 2.
Role of clock genes in leukemic transformation. The CLOCK-BMAL1 complex promotes the expression of clock genes and changes in the circadian clock. Furthermore, at high expression levels, CLOCK-BMAL1 can directly affect Wee1, which phosphorylates CDK1/cyclin B, interfering at the cell cycle transition from the G2 to the M phase. In addition, some circadian clock genes were reported at low levels, represented in yellow as PER, which altered the expression of cMyc, which is closely linked to cyclin D1. This also happens with NAPS2, which affects cMyc-Cyclin D1, leaving it overactivated. Cry1 presents a low-level expression that directly affects the expression of p21 and pro-inflammatory cytokines such as TNF-α and IL-6. Also at low expression levels, Cry2 can affect cMyc expression, which again supports its overactivity. In addition, Cry2 also affects p53 expression. Low levels of REV-ERBα and PPARα can also be correlated with changes in p21 expression. Furthermore, the REV-ERBα gene can also inhibit the cell cycle transition from the G1 to the S phase. The SHARP1 at high expression levels, as represented in green, has a correlation with the negative expression of CLOCK-BMAL1. The TIMELESS gene is also an important molecular biomarker, which—according to the studies—can present a high or low expression, participate in cell cycle checkpoints, and regulate Chk1 and Chk2, so it can cause an interruption in the G0 and G1 phases of the cell cycle. Finally, another gene that was shown to be at high and low levels was the CKIε gene, which—at its high levels—plays a role in the phosphorylation of the PER gene. Created with BioRender.com.