Abstract
Candida auris is considered a serious fungal pathogen frequently exhibiting a high resistance to a wide range of antifungals. In this study, a combination of the quorum-sensing molecule farnesol (FAR) and fluconazole (FLU) was tested on FLU-resistant C. auris isolates (C. auris S and C. auris R) compared to the susceptible C. auris H261. The aim was to assess the possible synergy between FAR and FLU, by reducing the FLU minimal inhibitory concentration, and to determine the mechanism underlying the conjunct effect. The results confirmed a synergic effect between FAR and FLU with a calculated FIC index of 0.75 and 0.4 for C. auris S and C. auris R, respectively. FAR modulates genes involved in azole resistance. When FAR was added to the cells in combination with FLU, a significant decrease in the expression of the CDR1 gene was observed in the resistant C. auris isolates. FAR seems to block the Cdr1 efflux pump triggering a restoration of the intracellular content of FLU. These results were supported by observed increasing accumulation of rhodamine 6G by C. auris cells. Moreover, C. auris treated with FAR showed an ERG11 gene down-regulation. Overall, these results suggest that FAR is an effective modulator of the Cdr1 efflux pump in C. auris and, in combination with FLU, enhances the activity of this azole, which might be a promising strategy to control infections caused by azole-resistant C. auris.
Keywords: Candida auris, antifungal resistance, farnesol, efflux pumps
1. Introduction
Finding an effective tool to combat the multidrug-resistant pathogen Candida auris is a challenge indeed. Isolated for the first time in 2009 in Japan, this fungus is currently widespread worldwide [1,2,3]. C. auris colonizes the skin and persists in the healthcare environment, frequently resulting in invasive outbreaks due to its capability to form adherent biofilms on clinically relevant surfaces [3,4,5]. Despite the increased incidence of C. auris infections, diagnostic tools remain difficult because of the lack of available conventional approaches. Additionally, therapeutic options are still limited [2,6]. C. auris isolates manifest resistance to common antifungals such as fluconazole (FLU) and amphotericin B (AMB) [7,8]. Moreover, resistance to echinocandins is emerging in some countries and has been shown to be acquired rather than intrinsic [4]. On the other hand, concomitant resistance to all three classes of antifungals (azoles, AMB, and echinocandins) is rare [3].
Overregulation of efflux pumps such as ATP binding cassette (ABC) and major facilitator superfamily (MFS) are known to cause multidrug resistance in Candida species [9]. Efflux together with mutations in the azole target gene ERG11 have been identified as the main mechanisms of resistance in C. auris [10,11]. Using a combination of antifungal drugs is a well-established approach for the treatment of several fungal infections, but to date, there is little evidence to support combined therapy in C. auris [12]. Searching for molecules able to modulate the efflux pumps synergizing with azoles against C. auris is a highly topical issue [13].
Farnesol (FAR) is a fungal quorum-sensing molecule that has been demonstrated in C. albicans to enhance the activity of antifungals and to affect the ABC efflux transporters resulting in decrease in the azole resistance [10,14]. The other favorable effect of FAR in combination with echinocandins (caspofungin and micafungin) was determined against Candida parapsilosis biofilms [15]. Another study showed the prominent effect of FAR with echinocandins against C. auris biofilms as well [16]. Furthermore, FAR blocked efflux pumps and down-regulated genes involved in biofilm development and resistance [5]. Modulation of C. auris efflux pump activity by FAR could represent a promising approach for controlling life-threatening infections caused by this pathogen [5,10]. Concerning obtained information, FAR should have a potential adjuvant effect in the eradication of resistant C. auris.
The aim of the presented study was to investigate the possible effect of the combination of FAR and FLU on the expression of the genes relevant to FLU resistance in C. auris strains manifesting different FLU susceptibility profiles.
2. Materials and Methods
2.1. Characterization of the C. auris Strains
Three isolates of C. auris were used in this study: C. auris H261 was kindly provided by Prof. Birgit Willinger (Medical University, Vienna, Austria). The strain was isolated in January 2018 from a healthy 22-year-old man with therapy-refractory external otitis. The detailed characterization, including the susceptibility profile, was published by Pekard-Amenitsch et al. (2018) [17]. The strain was deposited in the CBS-KNAW yeast collection of the Westerdijk Fungal Biodiversity Institute in Utrecht, the Netherlands, and assigned as CBS 15366. The isolates C. auris S and C. auris R were kindly provided by Prof. Maurizio Sanguinetti (Università Cattolica del Sacro Cuore, Rome, Italy). The clinical isolates were isolated from patients with positive blood cultures in hospitals in India. Strains were originally marked as C. auris 1a and C. auris 45 [18]. In the presented paper, the names have been changed due to different susceptibility profiles to FLU. The FLU-susceptible strain C. auris 1a was renamed as C. auris S and FLU-resistant strain C. auris 45 was renamed as C. auris R.
All strains were preserved at −80 °C in 1 mL of yeast extract peptone dextrose broth (YPD broth; 1% yeast extract, Biolife, Italy; 2% mycological peptone, Lab M Limited, Buri, UK; 2% D-glucose, Centralchem, Bratislava, Slovakia) supplemented with 60% glycerol (Centralchem, Bratislava, Slovakia). Prior to experiments, yeast cells from stocks stored at −80 °C were streaked onto a yeast extract–peptone–dextrose plate (YPD) supplemented with 2% agar (Biolife, Milan, Italy) and incubated overnight at 30 °C. One loopful of cells from YPD agar plates was inoculated into flasks containing 20 mL of YPD broth and grown in an orbital shaker (Multitrone Standard, Infors HT, Bottmingen-Basel, Switzerland), at 180 rpm for up to 16 h at 30 °C. The cells were then washed twice with phosphate-buffered saline (PBS) buffer (Sigma, St. Louis, MO, USA) and adjusted to the appropriate density for each experiment.
The susceptibility profile of C. auris isolates was determined by the broth microdilution method according to the European Committee on Antimicrobial Susceptibility Testing (EUCAST) protocol [19] in Roswell Park Memorial Institute medium (RPMI 1640 medium, Biowest, Nuaillé, France) supplemented with 2% D-glucose (Centralchem, Bratislava, Slovakia) and buffered with 0.165 M morpholinopropane sulfonic acid (MOPS, PanReac AppliChem, Darmstadt, Germany) to pH 7.0. Tested antifungals (fluconazole, FLU, Pfizer, New York, NY, USA; caspofungin, CAS, Merck Sharp & Dohme Ltd., London, UK; amphotericin B, AMB, Sigma Aldrich, Taufkirchen, Germany) were prepared in 2-fold serial dilutions ranging from 0.06–256 μg/mL for FLU, 0.015–16 μg/mL for CAS, and 0.06–8 μg/mL for AMB. Microtiter plates were incubated at 37 °C. After 24 h, optical density was measured at 570 nm using a plate reader (Dynex MRX-TC Revelation, Denkendorf, Germany). Susceptibility was evaluated in terms of minimal inhibitory concentration inhibiting the growth of the strain in the presence of the agent by 50% compared to the control sample without an antifungal agent (MIC50), and MIC90 representing the concentration that inhibits the growth of C. auris in the presence of AMB by 90% compared to the control sample without an agent. Each experiment was repeated at least 3 times with at least 4 parallel samples in each experiment.
2.2. Susceptibility Testing of FAR Alone and in Combination with FLU
Susceptibility testing was performed according to the EUCAST protocol as described above. The stock solution of 72 mM FAR (Sigma Aldrich, Taufkirchen, Germany) was prepared in 96% ethanol (Centralchem, Bratislava, Slovakia) and diluted to the following concentrations: 1 mM, 500 μM, 400 μM, 300 μM, 200 μM, 100 μM, 80 μM, 60 μM, and 50 μM in RPMI 1640 medium. The effectiveness of FAR was determined in terms of MIC50, as was previously described for antifungals. For determining the impact of FAR in combination with FLU, a subinhibitory concentration of FAR was tested (200 μM). Microtiter plates were seeded with 100 μL of the cell suspension (2 × 105 cells/mL) and 50 μL of FAR was added to wells, and the plate was incubated at 37 °C for 1 h. Finally, 50 μL of 2-fold serial dilutions of FLU (ranging from 0.06 to 256 μg/mL) was added to the wells. The microtiter plates were incubated at 37 °C for 24 h and OD was measured (Dynex MRX-TC Revelation, Denkendorf, Germany). The results were evaluated in terms of change in MIC50 of FLU in combination with FAR. Each experiment was repeated at least 3 times with at least 4 parallel samples in each experiment.
Drug–drug interactions were assessed by determining the fractional inhibitory concentration index (FICI) calculated according to the formula: FICI = FICI(A) + FICI(B), where FICI(A) = MIC(A) in combination/MIC(A) alone; FICI(B) = MIC(B) in combination/MIC(B) alone, where MIC(A) alone and MIC(B) alone are the MIC50 values of compounds A and B used alone and MIC(A) in combination and MIC(B) in combination are the MICs50 of compounds A and B at the effective combinations, respectively. The interaction between FLU and FAR was interpreted as synergistic when FICI was <0.5, as partially synergistic when FICI was ≥0.5 and <1, as an additive when FICI was 1, as indifferent when FICI was ≥ 1 and < 4, and as antagonistic when FICI was >4 [20,21].
2.3. qPCR Analyses of Genes Related to Resistance in the Presence of FLU and Combination FLU/FAR
At first, basal expression of genes related to the resistance (CDR1, CDR2, MDR1, and ERG11) was investigated by qPCR to determinate basal expression of tested genes. Briefly, total RNA was extracted from planktonic cells of C. auris harvested after overnight growth at YPD broth at 37 °C. Cells were washed twice with PBS and the pellet was prepared for isolation of RNA with GeneJET RNA Purification (ThermoScientific, Waltham, MA, USA) according to the manufacturer’s instructions. Then, we investigated the level of expression of the target genes after administration of FLU and FLU plus FAR. Total RNA was extracted as described above with slight modification in the yeast suspension preparation. C. auris isolates were grown overnight in YPD broth medium at 37 °C. After incubation, cells were centrifuged (5000× g for 5 min) and washed twice in sterile PBS and adjusted to concentration of 2 × 107 cells/mL in 10 mL of sterile YPD broth (a) without compounds (control); and supplemented with (b) FLU—subinhibitory concentrations (C. auris H261—0.06 μg/mL; C. auris S—8 μg/mL; C. auris R—32 μg/mL); (c) combination FAR and FLU: FAR (200 μM) at 37 °C for 1 h and afterwards with FLU (same concentration mentioned above) at 37 °C for an additional 1 h. Post incubation, cells were pelleted (5000× g for 5 min) and given a sterile PBS wash. The washed cells were re-suspended in sterile PBS (1 mL) and the suspension was used for RNA extraction. RNA samples were treated with DNase I, RNase-free (Thermo Scientific, Waltham, MA, USA) to prevent contamination with genomic DNA. The cDNA was synthesized using a Maxima First Strand cDNA Synthesis kit for RT-qPCR (Thermo Scientific, Waltham, MA, USA) according to the manufacturer’s instructions and stored at −20 °C until use. The PCR primers used to amplify and identify the C. auris genes CDR1, CDR2, MDR1, ERG11, as well as ACT1 genes are summarized in the Figure S2 and Table S1. All primers were synthesized by Metabion International AG (Planegg/Steinkirchen, Germany). For qPCR reaction, cDNA samples transcribed from 200 ng of total RNA were mixed with 5xHOT FIREPol® EvaGreen® qPCR Mix Plus (ROX) (Solis BioDyne, Tartu, Estonia) and amplified by using an Agilent Mx3000P qPCR System (Agilent Technologies, Inc., Santa Clara, CA, USA). Cycling conditions were as follows: one cycle of 15 min at 95 °C; followed by 40 cycles of 20 s at 95 °C and 30 s at 55 °C, followed by one cycle of 30 s at 72 °C for all genes. After amplification, a melting curve was run to ensure the absence of primer dimers.
The level of gene expression was calculated using the 2−ΔΔ CT method with respect to the housekeeping gene ACT1. Samples were compared to the control, which was represented by the most susceptible strain C. auris H261 cultivated without FLU or a combination FLU and FAR and normalized to 1. Each experiment was repeated at least three times with 3 parallel samples in each experiment.
2.4. Rhodamine 6G Intracellular Accumulation Assay and Fluorescence Microscopy
Intracellular accumulation assay was performed according to the protocol by Srivastava and Ahmad [5], with minor modification. Briefly, C. auris isolates were grown overnight in YPD broth medium at 37 °C. Then, C. auris suspensions were incubated with FLU and FAR as previously described in Section 2.3. Post incubation, cells were pelleted (5000× g for 5 min) and washed in sterile PBS. Cells were re-suspended in sterile PBS (1 mL) supplemented with 2% glucose and 4 μM rhodamine 6G (Sigma, Taufkirchen, Germany) and incubated at 37 °C for 30 min. Cells were then washed twice with cold sterile PBS and 1 mL of fresh PBS was added to the pellet. Afterwards, 100 μL of suspension was pipetted into a flat-bottomed dark 96 well plate (Costar®, Kennebunk, ME, USA) and fluorescence was measured with a fluorescence spectrophotometer (Tecan, Männedorf, Switzerland). The results were evaluated using MagellanTM Data Analysis Software and the intensity of fluorescence of samples was determined by relative fluorescence units (RFUs). The same suspension was immediately used for microscopy. Fluorescence was detected by an inverted fluorescence microscope (Zeiss, Jena, Germany) with excitation/emission spectra 525/548 nm. The pictures were captured by AxioCam ERc5s (Zeiss, Jena, Germany) and evaluated by software Motic Images Plus 3 (Hong Kong, China).
2.5. Statistical Analysis
Results were evaluated by statistical analysis using a one-way t-test in GraphPad Prism software (GraphPad, San Diego, CA, USA). Differences were considered statistically significant at p < 0.05 (*), p < 0.01 (**), p < 0.001 (***).
3. Results and Discussion
3.1. Identification of C. auris and Antifungal Susceptibility Profile of Tested Isolates
C. auris, also known as “the yeast superbug”, has become a serious emerging fungal pathogen since 2009 [1,22], but it might have emerged much earlier. Indeed, Candida haemuloni isolates from South Korea from 1996 have been re-identified and confirmed as C. auris, which suggested a high similarity between these two species [23]. Due to this observation, it is very important to use appropriate identification methods to correctly discriminate between C. auris and other Candida spp., mainly because of the increased resistance of this yeast to commonly used antifungals [22]. C. auris isolates from this study were controlled by phenotype and molecular analysis to avoid possible contamination (summarized in the Figure S1). The susceptibility profile was evaluated by determining the minimal inhibitory concentration (MIC) by a microdilution method to selected representatives of major classes of antifungals (FLU, CAS, and AMB). According to tentative breakpoints for C. auris published by the Center for Disease Control and Prevention (CDC; Atlanta, Georgia, USA) (https://www.cdc.gov/fungal/candida-auris/c-auris-antifungal.html, accessed on 29 May 2020), C. auris H261 manifested susceptibility to all tested antifungals (MIC50 rates: FLU = 0.125 μg/mL and CAS = 0.125 μg/mL; MIC90 rate: AMB = 0.5 μg/mL); C. auris S was susceptible to CAS (MIC50 = 0.125 μg/mL), resistant to AMB (MIC90 = 8 μg/mL), and showed decreased susceptibility to FLU (MIC50 for FLU = 16 μg/mL). C. auris R was resistant to all the three classes of antifungals (MIC50 rates: CAS = 8 μg/mL; FLU = 256 μg/mL, and MIC90 rate for AMB = 8 μg/mL). Our results agree with previously published data described for these C. auris isolates [17,18].
As was shown, the susceptibility profile of C. auris isolates used in this study was different and varied among isolates. Resistance to azoles has been reported in up to 91% of C. auris isolates [24]. In our study, C. auris S and C. auris R manifested a decreased susceptibility or resistance to FLU, whereas C. auris H261 remained susceptible to FLU as well as to both CAS and AMB. Susceptibility to azoles in C. auris is generally rare, but can be observed in healthy competent hosts that could be asymptomatically colonized with C. auris [17,25]. This is also the case of C. auris H261, which was isolated from the ear canal of an otherwise healthy man. On the other hand, azole resistance in C. auris remains a major problem in the treatment of candidiasis [9,24]. Some published works have already suggested that resistance could be associated with the regulation of genes involved in efflux or ergosterol biosynthesis [11,26]. Additionally, the resistance to multiple antifungal classes, as observed in C. auris R, is a phenomenon already reported [27,28,29,30,31,32].
3.2. Relative Expression Levels of Genes Related to the Azole Resistance (CDR1, CDR2, MDR1, and ERG11) in Untreated C. auris Isolates and after FLU and FAR Exposure
The overexpression of genes coding for the efflux pumps belonging to the ABC (CDR1, CDR2) and MFS (MDR1) superfamilies, together with the overexpression or point mutations of the ERG11 gene, are the main mechanisms responsible for azole resistance in Candida spp. [33]. The genome of C. auris contains genes coding for at least 20 transmembrane domains associated with ABC transporters that could potentially contribute to azole resistance [11].
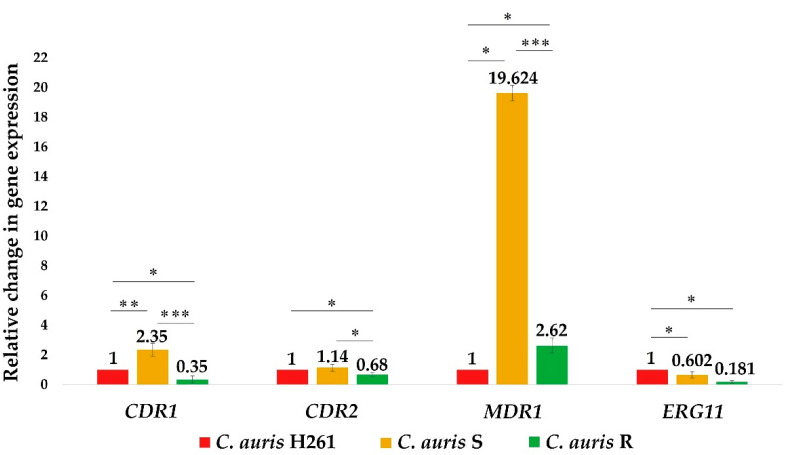
Expression of the above-mentioned genes (CDR1, CDR2, MDR1) and the ERG11 gene was analyzed in this study (Figure 1). Results showed a significantly increased regulation of the MDR1 gene in isolates of C. auris R and C. auris S (2.6 times and 19.6 in times increased, respectively), which was in agreement with previous observation indicating a resistance profile of both R and S isolates compared to the susceptible isolate H261. The efflux pump Mdr1p is a member of the MFS transporters responsible for the phenomenon of multiresistance in C. albicans [34,35]. Recently, Rybak et al. (2019) for the first time proved contribution of MDR1 and CDR1 to azole resistance in C. auris as well [26]. Our results agreed with this observation, confirming a native overregulation of the MDR1 gene in resistant C. auris isolates, despite the observation that the level of up-regulation of the MDR1 gene did not directly correspond to the level of the determined MIC50 to FLU in C. auris R. This indicated the participation of another mechanism of resistance against FLU as well.
Figure 1.
Basal expression of the CDR1, CDR2, MDR1, and ERG11 genes in C. auris isolates. The relative expression was calculated by qPCR using the 2−ΔΔ CT method with respect to the housekeeping gene ACT1 and using the susceptible isolate C. auris H261 for normalization. Data represent the average of 3 independent experiments performed in triplicate ± SD. p-values < 0.05 were considered significant: * <0.05; ** < 0.01; *** <0.001.
Interestingly, the level of native expression of the CDR genes was not significantly changed in bothl resistant isolates; the 2.3 times higher abundance was observed only in the case of C. auris S for the CDR1 gene. However, immediately after adding a subinhibitory concentration of FLU to both C. auris S and C. auris R, a significant up-regulation of the CDR1 gene was observed (Figure 4A). These results suggested an activation of the efflux pumps and confirmed the overexpression of the gene for the efflux pump Cdr1 as the major mechanism involved in C. auris resistance.
The expression of the ERG11 gene was down-regulated in both C. auris S and C. auris R (1.7 times and 5.5 times less, respectively) compared to the susceptible C. auris H261, suggesting possible changes in lipid homeostasis [36,37]. This down-regulation was particularly pronounced in C. auris R which could significantly contribute to resistance of this isolate. In a recent study, Shahi et al. (2020) performed a detailed lipidomic analysis of C. auris. The authors suggested that composition of lipids differs among isolates of C. auris and seems to affect their resistance profile. They found a lower content of intermediate compounds, such as squalene, lanosterol, fecosterol, episterol, and fungisterol formed in the earlier stage of the ergosterol pathway in resistant isolates [25]. This finding could support our observation of a decreased expression of the ERG11 gene in both C. auris R and S. Indeed, a down-regulation of the ERG 11 gene reflects a reduction in target protein production and can result in a decrease in the amount of lanosterol, as observed in the work of Shahi et al. (2020), that in turn can affect FLU efficacy [25]. Additionally, the down-regulation of the ERG11 gene in resistant isolates could also be triggered by mutations in this gene, another mechanism mediating resistance to azoles in C. auris [38,39].
3.3. The Synergy between FAR and FLU Modulates C. auris Resistance to FLU
The combination of FAR with common antifungals has already been described by many authors in C. albicans [14,40,41,42], but only a few studies discussed the potential use of FAR in C. auris [5,6,16]. The main part of this work was focused on the impact of the quorum-sensing molecule FAR on FLU-resistant C. auris isolates (C. auris S and C. auris R) compared to the susceptible isolate C. auris H261. Our results confirmed a partially synergistic effect between FAR and FLU, with a FIC index of 0.75 and 0.4 for C. auris S and C. auris R, respectively, that was supported by changes in expression of already mentioned resistance genes shown in the next paragraph. These results pointed to the possible role of FAR as a modulator of efflux pumps in resistant C. auris isolates.
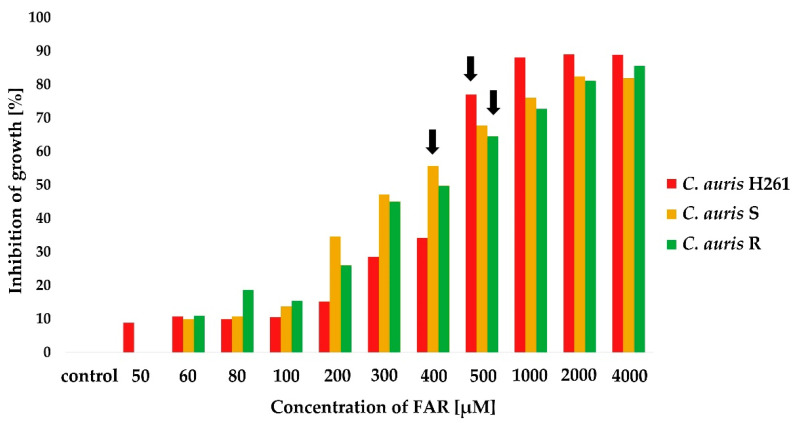
At first, MIC50 of FAR alone was tested by a microdilution method. The susceptibility of C. auris isolates was tested for 11 different concentrations of FAR. Results showed different rates of MIC50 for FAR among isolates (C. auris H261 = 500 μM, C. auris S = 400 μM, and C. auris R = 500 μM). Results are illustrated in Figure 2. Interestingly, MIC50 of FAR alone was generally lower in all the tested C. auris isolates compared C. albicans, which displayed a MIC50 for FAR of 1 mM [14]. This observation agreed with Nagy et al. (2020), who described the MIC50 of FAR to be slightly lower, i.e., 300 μM, for C. auris [6]. In addition, FAR susceptibility variations might be strain-dependent and could be influenced by the amount of intracellular FAR as well. As FAR is a substrate of the Cdr1 pump in C. albicans [43], we hypothesize that differences in the FAR MIC50 among tested C. auris isolates could rely on the level of the native expression of the CDR1 gene.
Figure 2.
Inhibitory effect of FAR on the planktonic cells of C. auris determined by MIC50 (labeled with black arrows). Optical density (OD580) of suspension was determined after 24 h cultivation of yeast in the presence of different concentrations of FAR; the control sample was without FAR. Percentage of growth inhibition was calculated from the OD580 values of samples compared to the inhibition of the control sample set to 0%. Data represent the average of 3 independent experiments performed in triplicate.
In the previous paragraph, the difference in expression of the genes related to resistance in C. auris isolates was described. Concretely, isolate C. auris S manifested the highest abundance of the CDR1 gene (2.35 times higher compared to C. auris H261) that could lead to a higher inhibitory effect of FAR alone on this isolate compared to C. auris H261 and C. auris R. With higher activity of the CDR1 gene, FAR could accumulate more in the cells, resulting in lower MIC for FAR in C. auris S, which was also found to be less susceptible to FLU. These results could be supported by Zamith-Miranda et al. (2018) who found higher expression of the gene coding for efflux transporter Cdr1 in FLU-resistant C. auris compared to FLU-susceptible C. albicans [37].
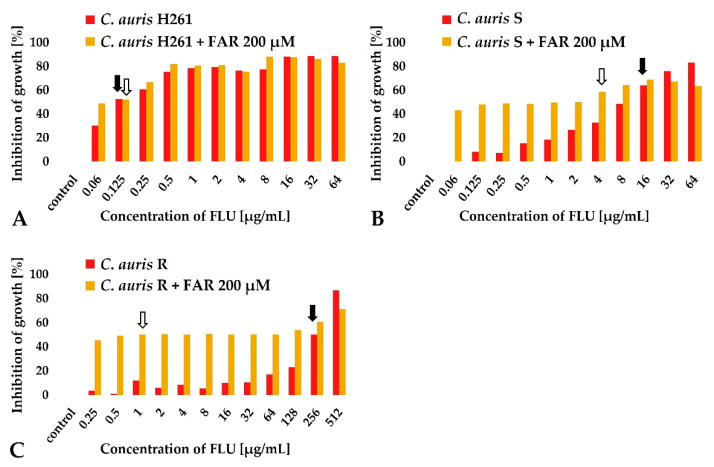
A subinhibitory concentration of FAR (200 μM) was selected to be tested in combination with FLU (Figure 3, gray columns). In the case of FLU-susceptible strain C. auris H261, FAR did not reduce FLU MIC50 values and the FICI was indifferent (Figure 3A). Changes in MIC50 were observed for both C. auris S (Figure 3B) and C. auris R (Figure 3C). A partial synergy was observed in C. auris S (the MIC50 for FLU decreased from 16 to 4 μg/mL), and a synergy was observed in C. auris R (the MIC50 for FLU decreased from 256 to 1 μg/mL). Results are summarized in Table 1. Our data are in agreement with the report of Nagy et al. (2020), confirming a synergism between FAR and voriconazole, posaconazole, itraconazole, and FLU, although the concentration of FAR used in the combination was lower (75 μM) compared to our data [6]. Interestingly, the same authors described a potential synergy of FAR in combination with echinocandins in C. auris [6].
Figure 3.
Synergistic effect of FAR (200 μM) in combination with FLU determined by changes in MIC50 of FLU. MIC50 values of FLU alone are marked with a black arrow. MIC50 of FLU combined with FAR are marked with a white arrow. MIC50 of FLU remained without change for C. auris H261 (A), MIC50 of FLU shifted from 16 μg/mL to 4 μg/mL for C. auris S (B), MIC50 of FLU shifted from 256 μg/mL to 1 μg/mL for C. auris R (C). Data represent the average of 3 independent experiments performed in triplicate.
Table 1.
Synergy between FLU and FAR (200 μM) of C. auris isolates determined according to MIC50 values.
| Strain | MIC50 FLU (Alone) (μg/mL) | Interpretation | MIC50 FAR (Alone) (μM) | MIC50 FLU with 200 μM FAR (μg/mL) | FIC Index | Interpretation |
|---|---|---|---|---|---|---|
| C. auris H261 | 0.125 | Susceptible | 500 | 0.125 | 1.4 | Indifferent |
| C. auris S | 16 | Susceptible * | 400 | 4 | 0.75 | Partial synergy |
| C. auris R | 256 | Resistant | 500 | 1 | 0.4 | Synergy |
FLU—Fluconazole; FAR—Farnesol; MIC50—Minimal inhibitory concentration of tested antifungals inhibiting the growth of yeast cells by 50% compared to the control sample without an agent. * Isolate C. auris S is susceptible according to the tentative breakpoint value for C. auris, which is 32 μg/mL (https://www.cdc.gov/fungal/candida-auris/c-auris-antifungal.html, accessed on 29 May 2020). However, MIC50 of 16 μg/mL is considered to be relatively high and means significantly decreased susceptibility of this isolate to FLU.
3.4. FAR Down-Regulates the Expression of Genes Involved in Resistance to FLU
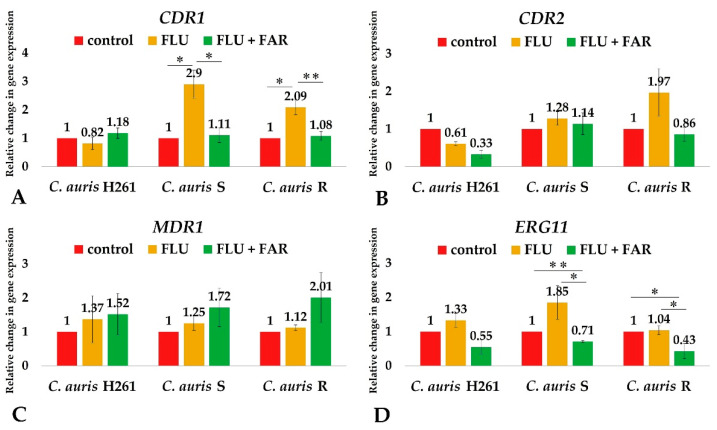
Recently published studies described the potential of FAR as an inhibitor of efflux pumps in resistant C. albicans [10,14]. Until now, only one report hypothesized a similar effect in C. auris [5], but the expression of genes was not investigated. qPCR was performed to investigate possible changes in the CDR1, CDR2, MDR1, and ERG11 expression after FAR plus FLU treatment in all the C. auris isolates.
We found an up-regulation of the CDR1 gene in both C. auris S and C. auris R (Figure 4A,B) treated with FLU. The relative fold change for CDR1 and CDR2 genes was higher in C. auris S (2.9 and 1.28 times for the CDR1 and CDR2 gene, respectively) and C. auris R (2.09 and 1.97 times for the CDR1 and CDR2 gene, respectively) compared to C. auris H261 (control). Our results agreed with the work of Wasi et al. (2019), proving that azole-resistant C. auris displays the overexpression of the CDR1 gene resulting in the efflux of FLU outside the cell [11]. On the contrary, we did not detect a significant change in the MDR1 expression after the addition of FLU to both resistant isolates of C. auris (Figure 4C). Accordingly, Hiller et al. (2006) showed a less efficient transport of FLU by Mdr1p in C. albicans [44]. In conclusion, our data support previous studies suggesting that a response of C. auris to the presence of FLU is mainly mediated by an efflux pump encoded by the CDR1 gene [7,9,11].
Figure 4.
Relative change expression of the CDR1 (A), CDR2 (B), MDR1 (C), and ERG11 (D) genes determined by qPCR. The expression of genes was calculated using the 2−ΔΔ CT method with respect to the housekeeping gene ACT1. The level of gene expression was examined in the presence of subinhibitory concentration of FLU: 0.06 μg/mL for C. auris H261, 8 μg/mL for C. auris S, and 32 μg/mL for C. auris R (the second column in each chart); a combination of 200 μM FAR and subinhibitory concentration of FLU (the third column in each chart). Values were compared to the control without any drugs (the first column in each chart) which was normalized to a value of 1. Data represent the average of 3 independent experiments performed in triplicate ± SD. p-values < 0.05 were considered significant: * < 0.05; ** < 0.01.
Afterwards, we studied changes in the gene expression after a combined treatment with FAR and FLU. A decrease in the expression of both the CDR1 and CDR2 genes was observed in resistant C. auris isolates (Figure 4A,B), suggesting an inhibitory effect of FAR on efflux transporters, but results proved to be significant only for the CDR1 gene.
The modulatory effect of FAR on the efflux pumps Cdr1 and Cdr2 was described for the first time in C. auris by Srivastava et al. in 2020 [5]. Our results agreed with that finding despite the concentration of FAR used in our experiments being much lower (200 μM) compared to that used by the mentioned authors (125 mM). We hypothesized that effectiveness of FAR on C. auris depends on up-regulation of the CDR1 gene. This claim was supported by observation that pointed to FAR’s ability to induce apoptosis in C. albicans that was mediated by the Cdr1 pump resulting in extrusion and depletion of intracellular glutathione [43,45]. In agreement with this conception, the basal expression of the CDR1 gene in C. auris S was higher compared to susceptible isolate C. auris H261. However, this was not confirmed in the most resistant isolate C. auris R. On the other hand, regardless of the basal data, FLU increased the CDR1 gene expression in both isolates. It is suggested that FLU might also activate the CDR1 gene in combination with FAR. Subsequently, FAR could effectively bind the Cdr1 pump to enter the cell, inhibit efflux, and help FLU be restored inside the cell. The presented results confirmed a synergy of FAR and FLU resulting in the increase in susceptibility of C. auris S and C. auris R resistant to FLU.
Additionally, a significant decrease in the ERG11 gene expression after administration of FAR in combination with FLU (Figure 4D) was observed in both C. auris S and C. auris R. The modulatory effect of FAR in the regulation of ergosterol synthesis has been described in C. albicans by Yu et al. (2012). Similarly, the down-regulation of the ERG1, ERG3, ERG6, ERG11, and ERG25 genes was observed in C. albicans biofilm treated with FAR [46]. The ERG25 gene (coding for methylsterol monooxygenase) and the ERG4 gene (coding for delta 24(24(1))-sterol reductase) were both shown to be down-regulated similarly to the FAR-exposed group of C. albicans described in the study of Wang and Liu (2019). It was also concluded that exogenous FAR has an evident, but non-deterministic, effect on the synthesis of ergosterol in C. albicans [47]. These results might suggest an indirect effect of FAR on ergosterol biosynthesis, although the mechanism underlying this ability is still largely unclear. Overexpression of the ERG11 gene is also known for playing a role in C. auris azole resistance [38,48]. The FAR-dependent down-regulation of the ERG11 gene in resistant strains of C. auris increased the inhibitory effect of FLU and could be a promising tool to overcome azole resistance.
Finally, changes in the MDR1 gene expression have not been observed in combination of FAR and FLU in resistant C. auris isolates (Figure 4C). From the point of view of FAR’s effect, the contribution of Mdr1 is not relevant in this study, due to the fact that FAR is not the substrate of this efflux transporter [43,45].
3.5. FAR Inhibits the Cdr1 Pump Allowing for a High Intracellular Accumulation of Rhodamine 6G
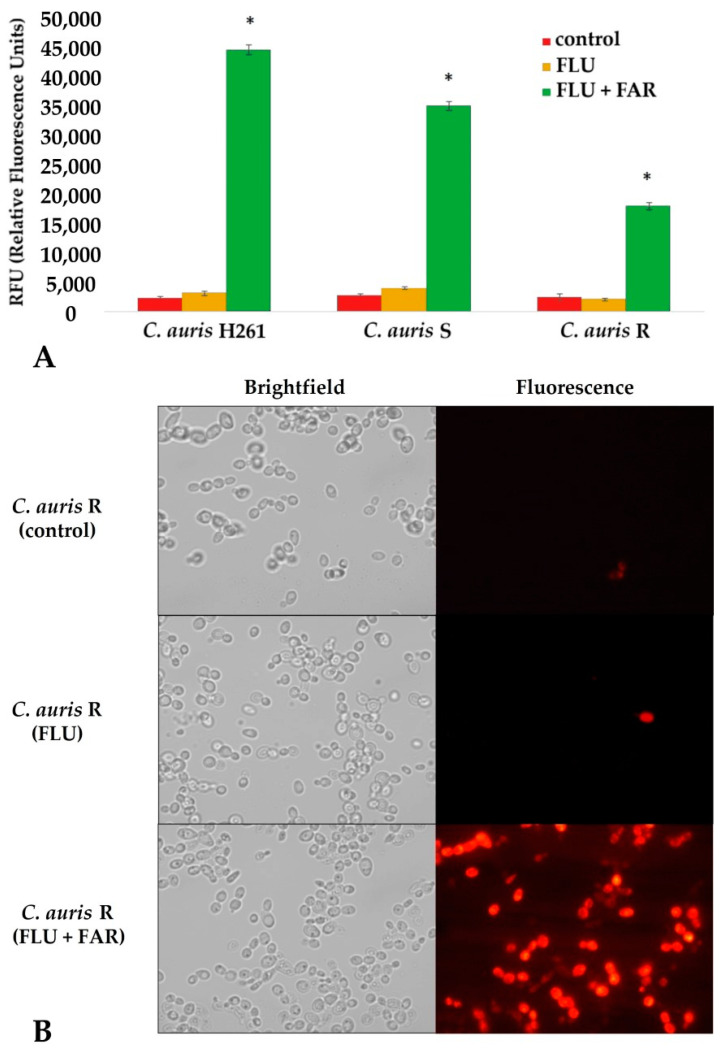
Results from the previous experiment suggested high activity of FAR as a potential modulator of the CDR1 and ERG11 gene expression related to azole resistance and the ergosterol pathway as well. To confirm the inhibitory effect of FAR on the activity of the C. auris Cdr1 efflux pump, the intracellular accumulation of rhodamine 6G dye (R6G) was studied in treated and untreated isolates. Untreated C. auris isolates were used as a control (Figure 5A, dark gray columns). FLU treatment alone resulted in a signal similar to that observed in the control group, confirming an active efflux (Figure 5A, light gray columns). The highest fluorescence signal has been observed in C. auris isolates treated with the FAR and FLU combination (Figure 5A, black columns). These results suggest a FAR-dependent inhibition of Cdr1 that blocks the efflux and restores the intracellular FLU. This hypothesis is supported by fluorescent microscopy. Indeed, there was a marked increase in the R6G fluorescence signal in cells treated with the combination of FAR and FLU compared to the untreated control or sample treated with FLU alone (Figure 5B, C. auris R). The same results were observed in both isolates C. auris H261 and C. auris S. These results corroborate qPCR data concerning the inhibitory effect of FAR on the CDR1 gene in combination with FLU. R6G is substrate for the Cdr1 pump in C. albicans, and it is very effective in the measurement of activity of this efflux transporter pump [49]. Srivastava et al. (2020) confirmed for the first time inhibition of Cdr1 with FAR in C. auris [5]. Our results are in agreement with that observation, but our study confirmed the effectivity of FAR and FLU synergy by molecular analysis. FAR in combination with FLU helps to accumulate FLU in the cells and blocks efflux through regulation of the CDR1 gene resulting in an increase in susceptibility of resistant C. auris isolates.
Figure 5.
(A) Rhodamine 6G intracellular accumulation measured by relative fluorescence. The figure shows a significant increase in the accumulation of R6G dye in all C. auris treated with a combination of FAR and FLU resulting in the highest abundance of fluorescence (green columns). The cells treated only with the subinhibitory concentration of FLU (yellow columns) have the same level of R6G as control cells without any agent (red columns) which means that R6G was extruded from cells by efflux. Subinhibitory concentrations of FLU were as follows: 0.06 μg/mL for C. auris H261, 8 μg/mL for C. auris S, and 32 μg/mL for C. auris R. (B) Fluorescence microscopy of the selected yeast suspension of the most resistant isolate C. auris R confirms the highest accumulation of R6G in the cells after a combination of FAR and FLU, confirming FAR as an inhibitor of efflux pump Cdr1; p-values * < 0.05 were considered significant.
4. Conclusions
Due to the strong impact of efflux in C. auris azole resistance, inhibitors/modulators of efflux pumps seem to represent a promising tool to overcome multiresistance of this yeast. FAR is a quorum-sensing molecule with a high potential to modulate efflux by inhibiting the Cdr1 transporter. Besides the inhibitory effect of FAR alone, the subinhibitory concentration of FAR in combination with FLU significantly decreased the expression of the CDR1 gene in agreement with a previously manifested shift of MIC50 for FLU. However, observed changes were shown to be C. auris strain-dependent, probably associated with a native expression of the studied genes. Additionally, the presented study proved for the first time that FAR contributed to decreased regulation of the ERG11 gene resulting in possible disturbance of lipid homeostasis. In summary, the presented data confirmed a promising role of FAR that can modulate resistance to azoles in resistant C. auris isolates and showed promising activity in combination with FLU.
Acknowledgments
The authors wish to thank Maurizio Sanguinetti for providing C. auris isolates.
Supplementary Materials
The following supporting information can be downloaded at: https://www.mdpi.com/article/10.3390/jof8080783/s1. Figure S1. Phenotype identification of the isolates of Candida sp. (A) C. albicans SC5314; (B) C. auris H261; (C) C. auris S; (D) C. auris R; Figure S2. Electrophoretogram of PCR products for genotype identification of C. auris and C. albicans; Table S1. List of oligonucleotide sequences used in this study. Reference [50] is cited in Supplementary file.
Author Contributions
Conceptualization—J.D., L.Č. and H.B.; methodology—J.D., S.K. and L.Č.; investigation—J.D., S.K., L.Č. and E.O.; writing—J.D., L.Č., S.K. and H.B.; review and editing—H.B., J.D., L.Č., E.B. and B.W. All authors have read and agreed to the published version of the manuscript.
Informed Consent Statement
Not applicable.
Conflicts of Interest
The authors declare no conflict of interest.
Funding Statement
This research was funded by the Slovak Research and Development Agency under contract APVV-15-0347 and APVV-18-0075 and by the grant VEGA 1/0537/19 supported by the Ministry of Education, Science, Research and Sport of the Slovak Republic.
Footnotes
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations.
References
- 1.Satoh K., Makimura K., Hasumi Y., Nishiyama Y., Uchida K., Yamaguchi H. Candida Auris Sp. Nov., a Novel Ascomycetous Yeast Isolated from the External Ear Canal of an Inpatient in a Japanese Hospital. Microbiol. Immunol. 2009;53:41–44. doi: 10.1111/j.1348-0421.2008.00083.x. [DOI] [PubMed] [Google Scholar]
- 2.Lone S.A., Ahmad A. Candida Auris-the Growing Menace to Global Health. Mycoses. 2019;62:620–637. doi: 10.1111/myc.12904. [DOI] [PubMed] [Google Scholar]
- 3.Iguchi S., Itakura Y., Yoshida A., Kamada K., Mizushima R., Arai Y., Uzawa Y., Kikuchi K. Candida Auris: A Pathogen Difficult to Identify, Treat, and Eradicate and Its Characteristics in Japanese Strains. J. Infect. Chemother. 2019;25:743–749. doi: 10.1016/j.jiac.2019.05.034. [DOI] [PubMed] [Google Scholar]
- 4.Lockhart S.R. Candida Auris and Multidrug Resistance: Defining the New Normal. Fungal Genet. Biol. 2019;131:103243. doi: 10.1016/j.fgb.2019.103243. [DOI] [PubMed] [Google Scholar]
- 5.Srivastava V., Ahmad A. Abrogation of Pathogenic Attributes in Drug Resistant Candida Auris Strains by Farnesol. PLoS ONE. 2020;15:e0233102. doi: 10.1371/journal.pone.0233102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Nagy F., Vitális E., Jakab Á., Borman A.M., Forgács L., Tóth Z., Majoros L., Kovács R. In Vitro and in Vivo Effect of Exogenous Farnesol Exposure Against Candida Auris. Front. Microbiol. 2020;11:957. doi: 10.3389/fmicb.2020.00957. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Kim S.H., Iyer K.R., Pardeshi L., Muñoz J.F., Robbins N., Cuomo C.A., Wong K.H., Cowen L.E. Genetic Analysis of Candida Auris Implicates Hsp90 in Morphogenesis and Azole Tolerance and Cdr1 in Azole Resistance. mBio. 2019;10:e02529-18. doi: 10.1128/mBio.02529-18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Romera D., Aguilera-Correa J.J., Gadea I., Viñuela-Sandoval L., García-Rodríguez J., Esteban J. Candida Auris: A Comparison between Planktonic and Biofilm Susceptibility to Antifungal Drugs. J. Med. Microbiol. 2019;68:1353–1358. doi: 10.1099/jmm.0.001036. [DOI] [PubMed] [Google Scholar]
- 9.Du H., Bing J., Hu T., Ennis C.L., Nobile C.J., Huang G. Candida Auris: Epidemiology, Biology, Antifungal Resistance, and Virulence. PLoS Pathog. 2020;16:e1008921. doi: 10.1371/journal.ppat.1008921. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Sharma M., Prasad R. The Quorum-Sensing Molecule Farnesol Is a Modulator of Drug Efflux Mediated by ABC Multidrug Transporters and Synergizes with Drugs in Candida Albicans. Antimicrob. Agents Chemother. 2011;55:4834–4843. doi: 10.1128/AAC.00344-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Wasi M., Khandelwal N.K., Moorhouse A.J., Nair R., Vishwakarma P., Bravo Ruiz G., Ross Z.K., Lorenz A., Rudramurthy S.M., Chakrabarti A., et al. ABC Transporter Genes Show Upregulated Expression in Drug-Resistant Clinical Isolates of Candida Auris: A Genome-Wide Characterization of ATP-Binding Cassette (ABC) Transporter Genes. Front. Microbiol. 2019;10:1445. doi: 10.3389/fmicb.2019.01445. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Bidaud A.L., Djenontin E., Botterel F., Chowdhary A., Dannaoui E. Colistin Interacts Synergistically with Echinocandins against Candida Auris. Int. J. Antimicrob. Agents. 2020;55:105901. doi: 10.1016/j.ijantimicag.2020.105901. [DOI] [PubMed] [Google Scholar]
- 13.Iyer K.R., Camara K., Daniel-Ivad M., Trilles R., Pimentel-Elardo S.M., Fossen J.L., Marchillo K., Liu Z., Singh S., Muñoz J.F., et al. An Oxindole Efflux Inhibitor Potentiates Azoles and Impairs Virulence in the Fungal Pathogen Candida Auris. Nat. Commun. 2020;11:6429. doi: 10.1038/s41467-020-20183-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Černáková L., Dižová S., Gášková D., Jančíková I., Bujdáková H. Impact of Farnesol as a Modulator of Efflux Pumps in a Fluconazole-Resistant Strain of Candida Albicans. Microb. Drug Resist. 2019;25:805–812. doi: 10.1089/mdr.2017.0332. [DOI] [PubMed] [Google Scholar]
- 15.Kovács R., Bozó A., Gesztelyi R., Domán M., Kardos G., Nagy F., Tóth Z., Majoros L. Effect of Caspofungin and Micafungin in Combination with Farnesol against Candida Parapsilosis Biofilms. Int. J. Antimicrob. Agents. 2016;47:304–310. doi: 10.1016/j.ijantimicag.2016.01.007. [DOI] [PubMed] [Google Scholar]
- 16.Nagy F., Tóth Z., Daróczi L., Székely A., Borman A.M., Majoros L., Kovács R. Farnesol Increases the Activity of Echinocandins against Candida Auris Biofilms. Med. Mycol. 2020;58:404–407. doi: 10.1093/mmy/myz057. [DOI] [PubMed] [Google Scholar]
- 17.Pekard-Amenitsch S., Schriebl A., Posawetz W., Willinger B., Kölli B., Buzina W. Isolation of Candida Auris from Ear of Otherwise Healthy Patient, Austria, 2018. Emerg. Infect. Dis. 2018;24:1596–1597. doi: 10.3201/eid2408.180495. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Orofino F., Truglio G.I., Fiorucci D., D’Agostino I., Borgini M., Poggialini F., Zamperini C., Dreassi E., Maccari L., Torelli R., et al. In Vitro Characterization, ADME Analysis, and Histological and Toxicological Evaluation of BM1, a Macrocyclic Amidinourea Active against Azole-Resistant Candida Strains. Int. J. Antimicrob. Agents. 2020;55:105865. doi: 10.1016/j.ijantimicag.2019.105865. [DOI] [PubMed] [Google Scholar]
- 19.EUCAST: Breakpoints for Antifungals Version 10.0, Valid from 2020-02-04. [(accessed on 4 May 2022)]. Available online: https://www.eucast.org/astoffungi/clinicalbreakpointsforantifungals/
- 20.Odds F.C. Synergy, Antagonism, and What the Chequerboard Puts between Them. J. Antimicrob. Chemother. 2003;52:1. doi: 10.1093/jac/dkg301. [DOI] [PubMed] [Google Scholar]
- 21.Dawis M.A., Isenberg H.D., France K.A., Jenkins S.G. In Vitro Activity of Gatifloxacin Alone and in Combination with Cefepime, Meropenem, Piperacillin and Gentamicin against Multidrug-Resistant Organisms. J. Antimicrob. Chemother. 2003;51:1203–1211. doi: 10.1093/jac/dkg238. [DOI] [PubMed] [Google Scholar]
- 22.Kean R., Brown J., Gulmez D., Ware A., Ramage G. Candida Auris: A Decade of Understanding of an Enigmatic Pathogenic Yeast. J. Fungi. 2020;6:30. doi: 10.3390/jof6010030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Lee W.G., Shin J.H., Uh Y., Kang M.G., Kim S.H., Park K.H., Jang H.-C. First Three Reported Cases of Nosocomial Fungemia Caused by Candida Auris. J. Clin. Microbiol. 2011;49:3139–3142. doi: 10.1128/JCM.00319-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Chen J., Tian S., Han X., Chu Y., Wang Q., Zhou B., Shang H. Is the Superbug Fungus Really so Scary? A Systematic Review and Meta-Analysis of Global Epidemiology and Mortality of Candida Auris. BMC Infect. Dis. 2020;20:827. doi: 10.1186/s12879-020-05543-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Shahi G., Kumar M., Kumari S., Rudramurthy S.M., Chakrabarti A., Gaur N.A., Singh A., Prasad R. A Detailed Lipidomic Study of Human Pathogenic Fungi Candida Auris. FEMS Yeast Res. 2020;20:foaa045. doi: 10.1093/femsyr/foaa045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Rybak J.M., Doorley L.A., Nishimoto A.T., Barker K.S., Palmer G.E., Rogers P.D. Abrogation of Triazole Resistance upon Deletion of CDR1 in a Clinical Isolate of Candida Auris. Antimicrob. Agents Chemother. 2019;63:e00057-19. doi: 10.1128/AAC.00057-19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Ademe M., Girma F. Candida Auris: From Multidrug Resistance to Pan-Resistant Strains. Infect. Drug Resist. 2020;13:1287–1294. doi: 10.2147/IDR.S249864. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Ostrowsky B., Greenko J., Adams E., Quinn M., O’Brien B., Chaturvedi V., Berkow E., Vallabhaneni S., Forsberg K., Chaturvedi S., et al. Candida Auris Isolates Resistant to Three Classes of Antifungal Medications—New York, 2019. MMWR Morb. Mortal. Wkly. Rep. 2020;69:6–9. doi: 10.15585/mmwr.mm6901a2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Fayed B., Jayakumar M.N., Soliman S.S.M. Caspofungin-Resistance in Candida Auris Is Cell Wall-Dependent Phenotype and Potential Prevention by Zinc Oxide Nanoparticles. Med. Mycol. 2021;59:1243–1256. doi: 10.1093/mmy/myab059. [DOI] [PubMed] [Google Scholar]
- 30.Sharma D., Paul R.A., Rudramurthy S.M., Kashyap N., Bhattacharya S., Soman R., Shankarnarayan S.A., Chavan D., Singh S., Das P., et al. Impact of FKS1 Genotype on Echinocandin In Vitro Susceptibility in Candida Auris and In Vivo Response in a Murine Model of Infection. Antimicrob. Agents Chemother. 2022;66:e0165221. doi: 10.1128/AAC.01652-21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Kathuria S., Singh P.K., Sharma C., Prakash A., Masih A., Kumar A., Meis J.F., Chowdhary A. Multidrug-Resistant Candida Auris Misidentified as Candida Haemulonii: Characterization by Matrix-Assisted Laser Desorption Ionization-Time of Flight Mass Spectrometry and DNA Sequencing and Its Antifungal Susceptibility Profile Variability by Vitek 2, CLSI Broth Microdilution, and Etest Method. J. Clin. Microbiol. 2015;53:1823–1830. doi: 10.1128/JCM.00367-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Escandón P., Chow N.A., Caceres D.H., Gade L., Berkow E.L., Armstrong P., Rivera S., Misas E., Duarte C., Moulton-Meissner H., et al. Molecular Epidemiology of Candida Auris in Colombia Reveals a Highly Related, Countrywide Colonization With Regional Patterns in Amphotericin B Resistance. Clin. Infect. Dis. 2019;68:15–21. doi: 10.1093/cid/ciy411. [DOI] [PubMed] [Google Scholar]
- 33.Whaley S.G., Berkow E.L., Rybak J.M., Nishimoto A.T., Barker K.S., Rogers P.D. Azole Antifungal Resistance in Candida Albicans and Emerging Non-Albicans Candida Species. Front. Microbiol. 2016;7:2173. doi: 10.3389/fmicb.2016.02173. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Wirsching S., Michel S., Köhler G., Morschhäuser J. Activation of the Multiple Drug Resistance Gene MDR1 in Fluconazole-Resistant, Clinical Candida Albicans Strains Is Caused by Mutations in a Trans-Regulatory Factor. J. Bacteriol. 2000;182:400–404. doi: 10.1128/JB.182.2.400-404.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Pourakbari B., Teymuri M., Mahmoudi S., Valian S.K., Movahedi Z., Eshaghi H., Mamishi S. Expression of Major Efflux Pumps in Fluconazole-Resistant Candida Albicans. Infect. Disord. Drug Targets. 2017;17:178–184. doi: 10.2174/1871526517666170531114335. [DOI] [PubMed] [Google Scholar]
- 36.Kean R., Ramage G. Combined Antifungal Resistance and Biofilm Tolerance: The Global Threat of Candida Auris. mSphere. 2019;4:e00458-19. doi: 10.1128/mSphere.00458-19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Zamith-Miranda D., Heyman H.M., Cleare L.G., Couvillion S.P., Clair G.C., Bredeweg E.L., Gacser A., Nimrichter L., Nakayasu E.S., Nosanchuk J.D. Multi-Omics Signature of Candida Auris, an Emerging and Multidrug-Resistant Pathogen. mSystems. 2019;4:e00257-19. doi: 10.1128/mSystems.00257-19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Healey K.R., Kordalewska M., Jiménez Ortigosa C., Singh A., Berrío I., Chowdhary A., Perlin D.S. Limited ERG11 Mutations Identified in Isolates of Candida Auris Directly Contribute to Reduced Azole Susceptibility. Antimicrob. Agents Chemother. 2018;62:e01427-18. doi: 10.1128/AAC.01427-18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.AlJindan R., AlEraky D.M., Mahmoud N., Abdalhamid B., Almustafa M., AbdulAzeez S., Borgio J.F. Drug Resistance-Associated Mutations in ERG11 of Multidrug-Resistant Candida Auris in a Tertiary Care Hospital of Eastern Saudi Arabia. J. Fungi. 2020;7:18. doi: 10.3390/jof7010018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Katragkou A., McCarthy M., Alexander E.L., Antachopoulos C., Meletiadis J., Jabra-Rizk M.A., Petraitis V., Roilides E., Walsh T.J. In Vitro Interactions between Farnesol and Fluconazole, Amphotericin B or Micafungin against Candida Albicans Biofilms. J. Antimicrob. Chemother. 2015;70:470–478. doi: 10.1093/jac/dku374. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Bozó A., Domán M., Majoros L., Kardos G., Varga I., Kovács R. The in Vitro and in Vivo Efficacy of Fluconazole in Combination with Farnesol against Candida Albicans Isolates Using a Murine Vulvovaginitis Model. J. Microbiol. 2016;54:753–760. doi: 10.1007/s12275-016-6298-y. [DOI] [PubMed] [Google Scholar]
- 42.Monteiro D.R., Arias L.S., Fernandes R.A., Deszo da Silva L.F., de Castilho M.O.V.F., da Rosa T.O., Vieira A.P.M., Straioto F.G., Barbosa D.B., Delbem A.C.B. Antifungal Activity of Tyrosol and Farnesol Used in Combination against Candida Species in the Planktonic State or Forming Biofilms. J. Appl. Microbiol. 2017;123:392–400. doi: 10.1111/jam.13513. [DOI] [PubMed] [Google Scholar]
- 43.Zhu J., Krom B.P., Sanglard D., Intapa C., Dawson C.C., Peters B.M., Shirtliff M.E., Jabra-Rizk M.A. Farnesol-Induced Apoptosis in Candida Albicans Is Mediated by Cdr1-p Extrusion and Depletion of Intracellular Glutathione. PLoS ONE. 2011;6:e28830. doi: 10.1371/journal.pone.0028830. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Hiller D., Sanglard D., Morschhäuser J. Overexpression of the MDR1 Gene Is Sufficient to Confer Increased Resistance to Toxic Compounds in Candida Albicans. Antimicrob. Agents Chemother. 2006;50:1365–1371. doi: 10.1128/AAC.50.4.1365-1371.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Kong E.F., Tsui C., Kucharíková S., Van Dijck P., Jabra-Rizk M.A. Modulation of Staphylococcus Aureus Response to Antimicrobials by the Candida Albicans Quorum Sensing Molecule Farnesol. Antimicrob. Agents Chemother. 2017;61:e01573-17. doi: 10.1128/AAC.01573-17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Yu L., Wei X., Ma M., Chen X., Xu S. Possible Inhibitory Molecular Mechanism of Farnesol on the Development of Fluconazole Resistance in Candida Albicans Biofilm. Antimicrob. Agents Chemother. 2012;56:770–775. doi: 10.1128/AAC.05290-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Wang F.-J., Liu Z.-H. Systematic Analysis of Protein Expression in Candida Albicans Exposed to Farnesol. Chin. Med. J. Engl. 2019;132:2348–2353. doi: 10.1097/CM9.0000000000000420. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Bhattacharya S., Holowka T., Orner E.P., Fries B.C. Gene Duplication Associated with Increased Fluconazole Tolerance in Candida Auris Cells of Advanced Generational Age. Sci. Rep. 2019;9:5052. doi: 10.1038/s41598-019-41513-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Maesaki S., Marichal P., Vanden Bossche H., Sanglard D., Kohno S. Rhodamine 6G Efflux for the Detection of CDR1-Overexpressing Azole-Resistant Candida Albicans Strains. J. Antimicrob. Chemother. 1999;44:27–31. doi: 10.1093/jac/44.1.27. [DOI] [PubMed] [Google Scholar]
- 50.Ruiz-Gaitán A.C., Fernández-Pereira J., Valentin E., Tormo-Mas M.A., Eraso E., Pemán J., de Groot P.W.J. Molecular identification of Candida auris by PCR amplification of species-specific GPI protein-encoding genes. Int. J. Med. Microbiol. 2018;308:812–818. doi: 10.1016/j.ijmm.2018.06.014. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.