Figure 1. Amino acids in the N terminus of E3 that impart Z-NA binding are required to restrict VACV-induced necroptosis.
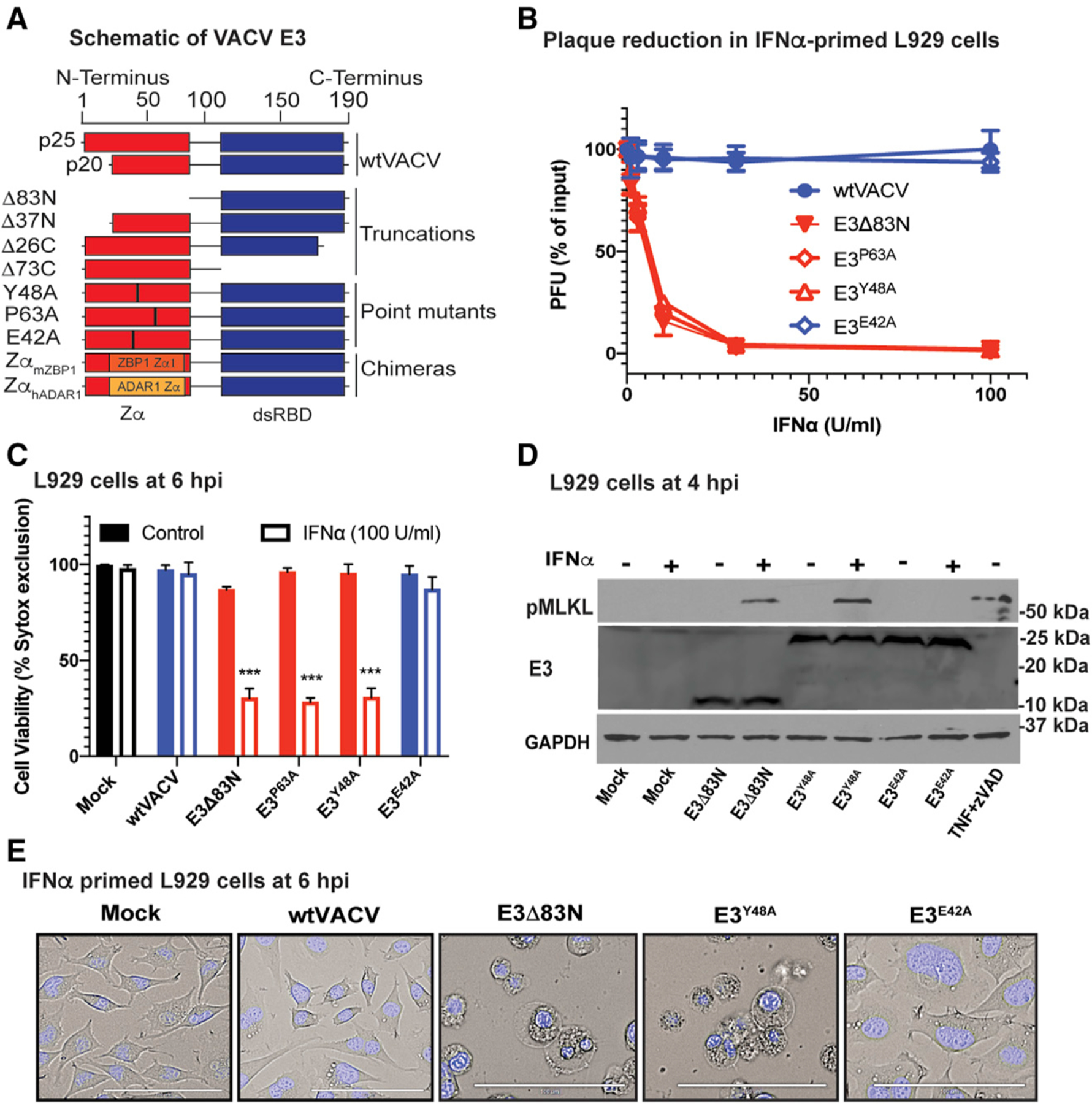
(A) A schematic diagram of VACV E3 and E3 derivatives made previously (Kim et al., 2003) and used in this study, including truncation mutants (E3Δ83N, E3Δ37N, E3Δ26C, and E3Δ73C), single amino acid substitution mutants (E3Y48A, E3P63A, and E3E42A) and chimeric E3 with Zα substitutions (ZαhADAR1 and ZαmZBP1).
(B) Plaque reduction assay performed by pretreating L929 cells with increasing concentrations of mouse IFN-α for 18 h prior to infection. Percent reduction of PFU was calculated as a proportion of input PFU for each virus evaluated. E3Y48A and E3P63A point mutations disrupt Z-NA binding; whereas, E3E42A mutation is at a non-essential location of the Zα so this protein retains Z-NA binding and is included as a control (Kim et al., 2003).
(C) L929 cells either left untreated or treated with mouse IFN-α (100 U/mL) for 18 h and subsequently infected at a MOI of 5 with each of the viruses shown. Cell viability was determined by SYTOX dye exclusion by counting an average of 10 fields microscopically at 6 hpi in three individual experiments.
(D)Immunoblot (IB) of L929 cell lysates either left untreated (−) or pretreated (+) with IFN-α and prepared at 4 hpi (MOI of 5 with indicated viruses) for phospho-MLKL, total E3 levels and GAPDH (as a loading control) following separation by SDS-PAGE. Mock infected cells and cells induced into necroptosis by TNF at 25 ng/mL in the presence of 50 µM zVAD-fmk from 1 h prior to TNF (Koehler et al., 2017) are included as a negative and positive controls, respectively.
(E) Phase contrast micrographs showing the morphology of L929 cells pretreated with IFN-α and left uninfected (mock) or infected at a MOI of 5 with the indicated viruses before being stained at 6 hpi with cell-permeant Hoechst 33342 and visualized. Bar indicates 100 µM.
Error bars represent the standard deviation (SD). Figures are representative of three independent replicates, except (B) and (C), which compiles the results of the replicates. Statistical significance was determined by a nonparametric Mann-Whitney test using GraphPad Prism9 with p ≤ 0.05 considered significant (*p ≤ 0.05, **p ≤ 0.01, ***p ≤ 0.001).
