Figure 6. Equivalence of Z-NA binding family Zα substitutions in suppressing necroptosis.
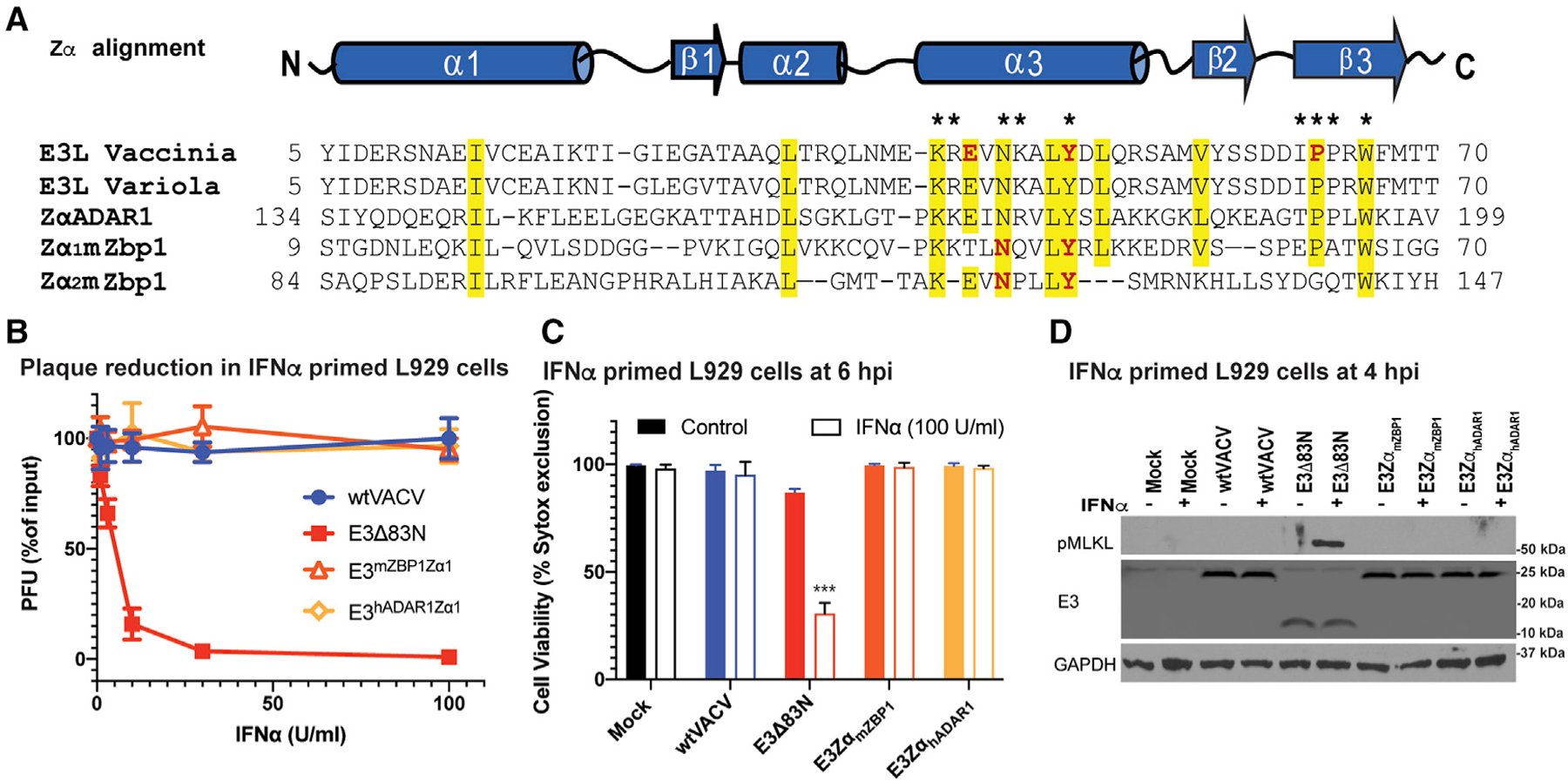
(A) Sequence alignment of the Za of VACV E3, variola E3, hADAR1 and mouse Zα1 and Zα2. Yellow signifies >70% sequence identity, amino acids with an asterisk above are known to be required for ZNA binding, and those in red have been targeted by point mutations (Kim et al., 2004, 2003). The GenBank accession numbers for the various sequences are as follows: GenBank: AAA02759 (vaccinia virus); GenBank: NP_042088 (variola virus); ADAR 1 (Homo sapiens): GenBank: AAB06697; ZBP1 (Mus musculus): GenBank: NP_067369.
(B) Plaque reduction assays using Zα chimeric viruses were performed on IFNα-primed L929 cells as described in Figure 1.
(C) L929 cells left untreated or treated with IFNα for 18 h and subsequently infected at a MOI of 5 with the indicated viruses. Cell viability was determined at 6 hpi using by Sytox dye exclusion as described in Figure 1.
(D) IB of L929 cell lysates from cells either left untreated or IFN-α-pretreated as described in Figure 1 and then infected with a MOI of 5 with the indicated viruses. Lysates were harvested at 4 hpi, subjected to SDS-PAGE and evaluated for phospho-MLKL.
Error bars represent the SD. Statistical significance was determined as described in Figure 1. Each set of data is representative of three replicates except for (B) and(C), which compiles the results of the replicates. Statistical significance was determined as described in Figure 1.
