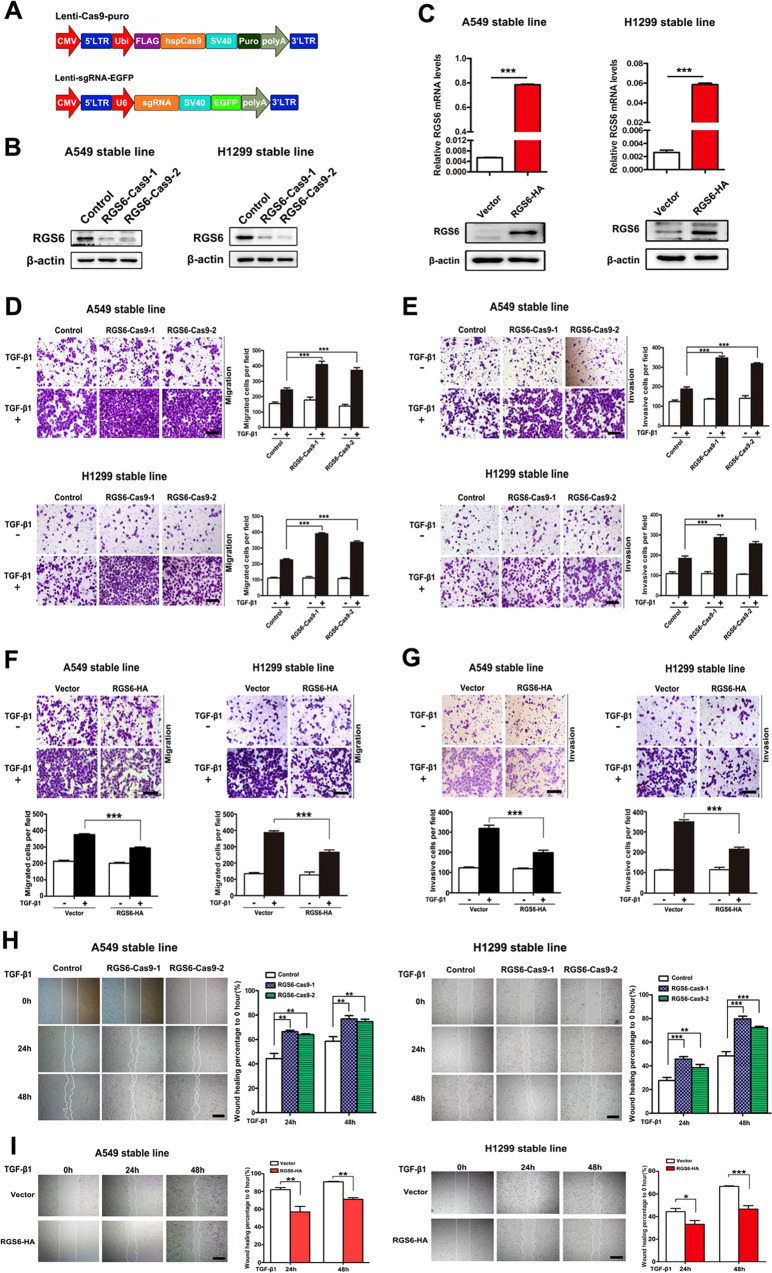
Fig. 2. Loss of RGS6 boots TGF-β-induced EMT events in NSCLC cells, whereas overexpression of RGS6 represses TGF-β-induced EMT events NSCLC cells.
A Schematic diagram of the CRISPR/Cas9 lentiviral system. B Confirmation of generation of two RGS6-KO NSCLC cell lines by western blotting. C Generation of NSCLC cell lines stably overexpressing RGS6. qRT-PCR (upper) and western blotting (lower) were used to confirm RGS6 overexpression. D, F In the presence or absence of TGF-β, indicated cells were allowed to migrate through 8-μm pores on the bottom of the transwell insert for 24 h. Migrated cells were stained and counted in at least three microscopic fields. Representative images (D left, F upper) and quantitated data (D right, F lower) are shown. Scale bar, 100 μm (***p < 0.001). E, G In the presence or absence of TGF-β, indicated cells were allowed to invade through the Matrigel-coated transwell insert for 24 h. Invasive cells were stained and counted in at least three microscopic fields. Representative images (E left, G upper) and quantitated data (E right, G lower) are shown. Scale bar, 100 μm. (**p < 0.01, ***p < 0.001) H, I Wound-healing assay was performed to evaluate TGF-β-induced (5 ng/ml) cell migration of indicated cells. The wound edges were photographed at the indicated time points after wounding. Each experiment was performed in triplicates. Representative images (left) and representative quantitated data (right) are shown. Scale bar, 200 μm. (**p < 0.01, ***p < 0.001).