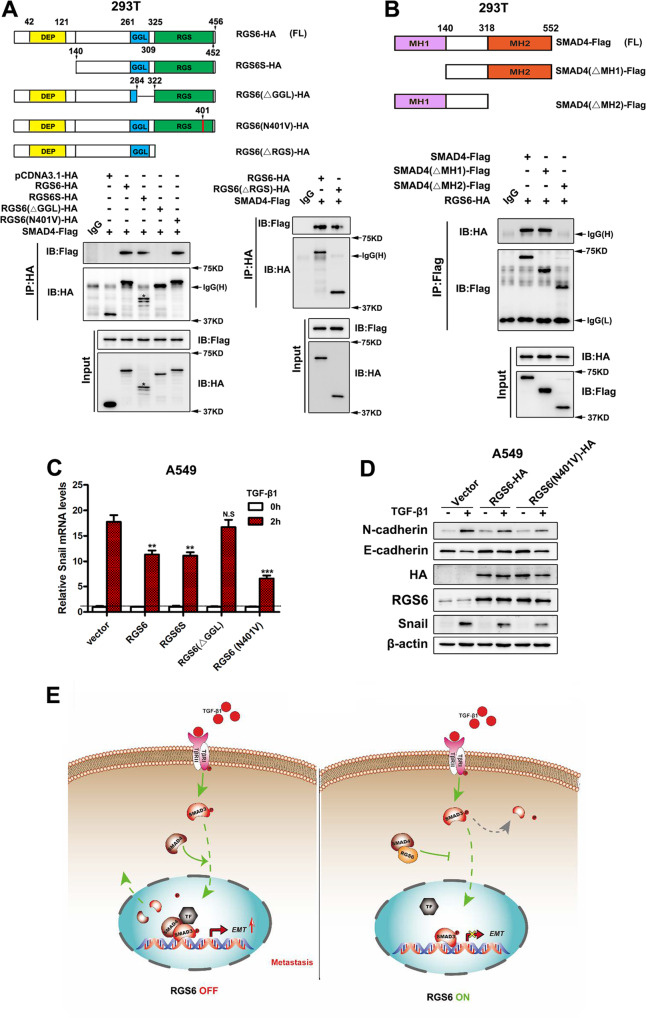
Fig. 7. Interaction between RGS6 and SMAD4 is mediated via association between the GGL domain and the MH2 domain.
A Upper, schematic illustration of RGS6 mutants. Lower, 293 T cells were co-transfected with Flag-tagged SMAD4 and various HA-tagged RGS6 mutants. Cell lysates were collected and subjected to IP with anti-HA antibody and probed for indicated proteins. B Upper, schematic illustration of SMAD4 mutants. Lower, 293 T cells were co-transfected with HA-tagged RGS6 and various Flag-tagged SMAD4 mutants. Cell lysates were collected and subjected to IP with anti-Flag antibody and probed for indicated proteins. C A549 cells transfected with different RGS6 mutants were treated with or without TGF-β (5 ng/ml) for 2 h. Cell lysates were collected and subjected to qRT-PCR for detection of Snail mRNA level (**p < 0.01, ***p < 0.001). D A549 cells transfected with different RGS6 mutants were treated with or without TGF-β (5 ng/ml) for 24 h. Cell lysates were collected and subjected to immunoblotting for indicated proteins. E Schematic diagram summarizing the effects of RGS6 on the TGF-β-SMAD pro-EMT signaling. Upon TGF-β stimulation, complex formation between SMAD4 and phosphorylated R-SMAD, such as SMAD3, facilitates nuclear translocation of p-SMAD3 into the nucleus, where SMAD4 further promotes association between p-SMAD3 and co-activators (TF) to ensure full efficiency of SMAD3-mediated gene expression, followed by dephosphorylation of SMAD3 and recycling of SMAD3 and SMAD4 back into the cytoplasm. RGS6 interacts with SMAD4 and prevents complex formation between SMAD4 and p-SMAD3. Interaction between RGS6 and SMAD4 causes cytoplasmic retaining of p-SMAD3 and SMAD4, leading to less p-SMAD3 in the nucleus and poor association between p-SMAD3 and co-activators, finally resulting in inefficient SMAD3-mediated gene expression.