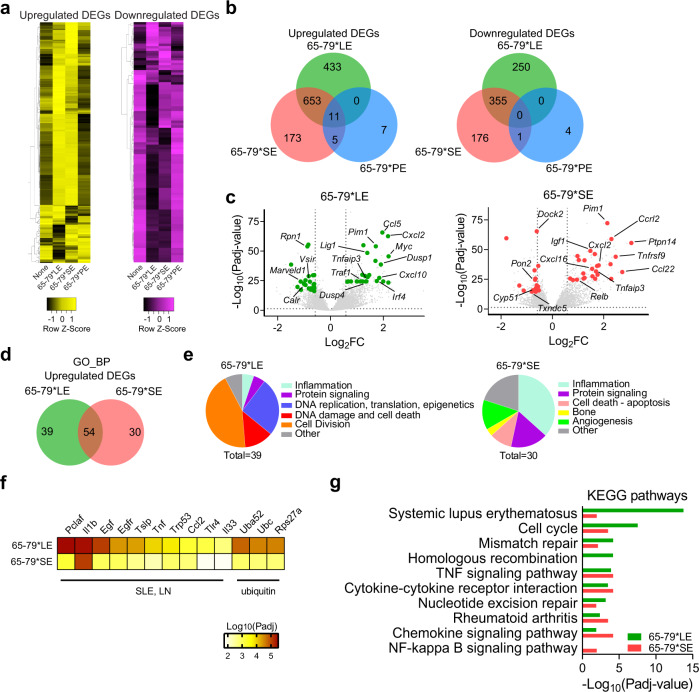
Fig. 1. Transcriptional modulation by the LE (Model A: epitope + IFN-γ, versus IFN-γ alone).
a Heatmaps showing unsupervised clustering of upregulated (left, yellow) and downregulated (right, purple) DEGs for 65-79*LE, 65-79*SE and 65-79*PE. DEGs were selected based on a > 3-fold change for 65-79*LE. All depicted DEGs had a Padj < 0.05. b Venn diagrams showing comparison of DEGs for 65-79*LE, 65-79*SE and 65-79*PE. c Volcano plots comparing transcriptional modulation by 65-79*LE and 65-79*SE. Green dots denote SLE-relevant genes modulated by 65-79*LE; red dots denote RA-relevant genes modulated by 65-79*SE. d Venn diagram comparing GO-BP terms enrichment among upregulated DEGs for 65-79*LE versus 65-79*SE. e Pie charts showing differential thematic clustering of unique GO-BP term enrichment among 65-79*LE- (left) versus 65-79*SE- (right) upregulated DEGs. f Heatmap of top-ranked SLE- or ubiquitination-associated URs that are predicted to be involved in 65-79*LE transcriptional modulation, as compared to their predicted involvement in 65-79*SE transcriptional modulation. g Notable KEGG pathway enrichment among 65-79*LE-upregulated DEGs, versus 65-79*SE. Data in (b–g) are based on DEGs with a fold change >1.5 and Padj < 0.05. All data are based on 6 biological replicates in 2 independent experiments. See also Supplementary Tables 1 and 4.