Significance
Infections with methicillin-resistant Staphylococcus aureus (MRSA) are associated with significant morbidity and mortality. Vancomycin is a last-line antibiotic used to treat MRSA infections; however, strains with decreased susceptibility to vancomycin (vancomycin-intermediate S. aureus [VISA]) have been spreading, and VISA infections are associated with prolonged therapeutic treatment and treatment failure. To map out the evolutionary trajectory behind VISA development, we characterized the mutational, transcriptional, and phenotypic landscape of 10 lineages of S. aureus USA300 strain JE2 that evolved in parallel to vancomycin. We demonstrate that MRSA strains adapt to vancomycin by divergent pathways leading to high or low oxacillin susceptibility characterized by mutational or transcriptional profiles. Our results point to diagnostic possibilities that may support personalized antibiotic treatment regimes.
Keywords: antibiotic resistance, adaptive laboratory evolution, transcriptional regulation, virulence
Abstract
Human infections with methicillin-resistant Staphylococcus aureus (MRSA) are commonly treated with vancomycin, and strains with decreased susceptibility, designated as vancomycin-intermediate S. aureus (VISA), are associated with treatment failure. Here, we profiled the phenotypic, mutational, and transcriptional landscape of 10 VISA strains adapted by laboratory evolution from one common MRSA ancestor, the USA300 strain JE2. Using functional and independent component analysis, we found that: 1) despite the common genetic background and environmental conditions, the mutational landscape diverged between evolved strains and included mutations previously associated with vancomycin resistance (in vraT, graS, vraFG, walKR, and rpoBCD) as well as novel adaptive mutations (SAUSA300_RS04225, ssaA, pitAR, and sagB); 2) the first wave of mutations affected transcriptional regulators and the second affected genes involved in membrane biosynthesis; 3) expression profiles were predominantly strain-specific except for sceD and lukG, which were the only two genes significantly differentially expressed in all clones; 4) three independent virulence systems (φSa3, SaeR, and T7SS) featured as the most transcriptionally perturbed gene sets across clones; 5) there was a striking variation in oxacillin susceptibility across the evolved lineages (from a 10-fold increase to a 63-fold decrease) that also arose in clinical MRSA isolates exposed to vancomycin and correlated with susceptibility to teichoic acid inhibitors; and 6) constitutive expression of the VraR regulon explained cross-susceptibility, while mutations in walK were associated with cross-resistance. Our results show that adaptation to vancomycin involves a surprising breadth of mutational and transcriptional pathways that affect antibiotic susceptibility and possibly the clinical outcome of infections.
Vancomycin-intermediate Staphylococcus aureus (VISA) strains emerge following the stepwise accumulation of mutations conferring low increments in resistance levels, and they display vancomycin minimal inhibitory concentrations (MIC) of 4 to 8 µg/mL compared to 2 µg/mL or less for the vancomycin-sensitive strains (VSSA) (1). Evolution to VISA status occurs upon exposure to vancomycin or other cell wall active antibiotics including β-lactams (2) and commonly involves a state of heteroVISA (hVISA) in which the bacterial population appears susceptible but subpopulations of cells display resistance (3).
Adaptation to vancomycin has been studied in both adaptive laboratory experiments (ALEs) and the clinic, with strains retrieved from patients before and after treatment (4–7). The results reveal a significant overlap in the genetic changes observed in vivo and in vitro, and the mutations affect major cellular processes including transcription, metabolism, and cell wall biosynthesis (8). Particularly affected are several two- and three-component signal transduction systems. These include the vancomycin resistance–associated VraT(YvqF)/VraSR three-component system that controls the expression of central peptidoglycan biosynthesis products (9), the GraSR system that together with GraX and the VraFG ATP-binding cassette transporter forms a five-component system that controls lysylination and alanylation of the cell envelope (10), and the essential WalKR system that regulates peptidoglycan hydrolase activity (11).
To determine the mutational pathways leading to VISA, we deployed ALE to evolve a community-associated S. aureus strain, JE2, to vancomycin stress. We sequenced lineages across two timepoints and surveyed the functional systems affected. The complexity of mutations and transcriptional changes associated with decreased vancomycin susceptibility prompted us to predict that VISA strains may be characterized by common transcriptional signatures. To our surprise, we found little obvious commonality between the transcriptional responses of VISA strains obtained by ALE to vancomycin despite fixed evolution settings and a common ancestor. Therefore, we turned to independent component analysis (ICA) to dissect transcriptomic datasets into groups of genes with independently modulated transcriptional signals, termed iModulons (12). Our results pointed to five iModulons characterizing VISA strains: φSa3, SaeR, T7SS, VraR, and Rex/NreC. Notably, we also observed that adaptation to vancomycin took at least two paths, leading to either high (> 256 μg/mL) or low (< 1 μg/mL) resistance to oxacillin compared to 32 μg/mL for the JE2 strain used for the evolution experiment. This divergence is related to specific activity within these iModulons and correlated with increased susceptibility to inhibitors of either cell wall or lipoteichoic acid synthesis. Our analysis demonstrates the power of ICA in dissecting complex regulatory responses and points to previously unrecognized regulators as contributors to the VISA phenotype.
Results
Parallel adaptation of S. aureus USA300 strain JE2 yields phenotypically diverse lineages reminiscent of clinical VISA strains.
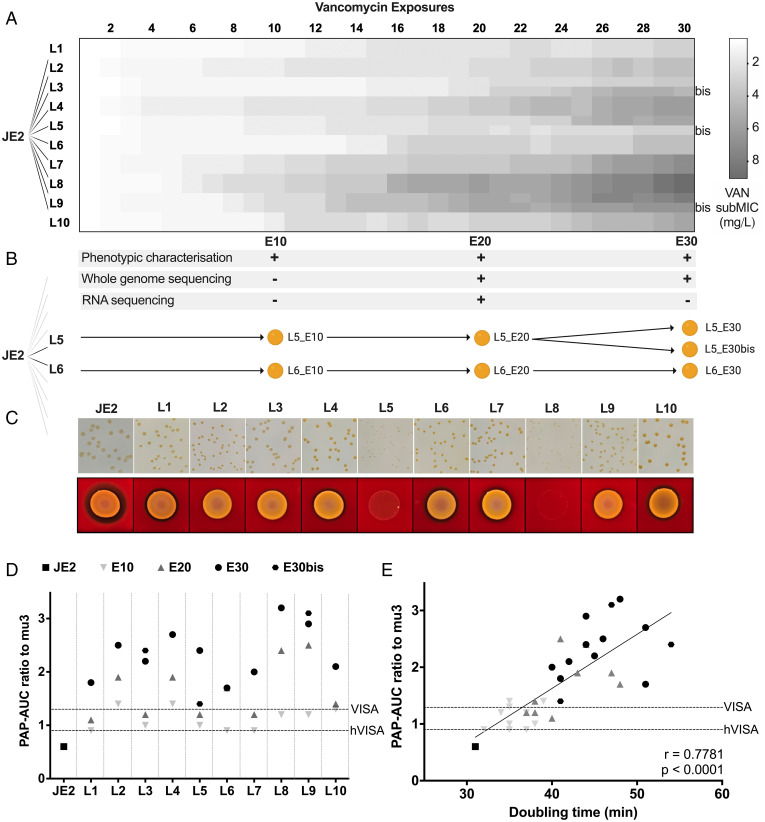
To phenotypically characterize the adaptation of a vancomycin-susceptible strain, we challenged the USA300 MRSA strain JE2 with increasing concentrations of vancomycin in 10 parallel cultures for 30 cycles of vancomycin exposure (E). Aliquots of the 10 independent lineages (L1–L10) were saved at each cycle to recount the chronological evolution from VSSA to VISA and were characterized in detail at three distinct timepoints: E10, E20, and E30 (Fig. 1 A and B). For technical reasons, three lineages were duplicated at E20 and grown as independent cultures until E30, resulting in the additional clones L3_E30bis, L5_E30bis, and L9_E30bis. Susceptibility testing was conducted using broth dilution (Fig. 1A) and population analysis profile-area under the curve (PAP-AUC) was performed by plating culture dilutions on increasing antibiotic concentrations and recording the number of colonies formed (13) (Fig. 1D).
Fig. 1.
Adaptive evolution of MRSA strain JE2 to increasing concentrations of vancomycin (VAN) and phenotypic changes. (A) JE2 was grown in TSB on increasing concentrations of vancomycin for 30 exposure cycles in 10 independent parallel lineages using a serial transfer protocol (see Methods). Shades of gray indicate vancomycin subMIC ranging from 0.5 μg/mL (white) to 9 μg/mL (gray). (B) Single clones from the 10 lineages were characterized at timepoints E10, E20, and E30 at the phenotypic and/or genetic and/or transcriptional level. Some lineages were split into two independent evolutions between E20 and E30, resulting in additional clones L3_E30bis, L5_E30bis, and L9_E30bis. (C) Colony morphology and blood hemolysis of clones at E20. (D) Vancomycin susceptibility of JE2-derived VISA clones at E10, E20, and E30 compared to wild type by PAP-AUC. (E) Scatterplot of the doubling time of the evolved clones against the ratio of measured vancomycin PAP-AUC with respect to wild type. Horizontal dashed lines represent the susceptibility threshold characterizing hVISA and VISA strains.
By the end of the evolution experiment, all lineages of JE2 reached VISA status with subMICs (i.e., the highest concentration at which growth was observed) increasing from 0.5 μg/mL at the start of the experiment to between 3 and 9 μg/mL at E30 (Fig. 1A). There was substantial variation in the time it took for each lineage to increase its MIC, and the increases required small increments in vancomycin concentration that rarely exceeded 0.5 μg/mL. In addition, we observed large variations in phenotypes of the resulting clones in terms of colony size, degree of pigmentation, and hemolysis (Fig. 1C). While these phenotypes did not correlate with vancomycin susceptibility, there was an inverse correlation with growth rate (Fig. 1E and SI Appendix, Table S1). Decreased hemolysis and fitness have previously been observed for VISA strains (14). Overall, VISA strains appear phenotypically diverse despite originating from a common ancestor.
Several disparate mutational trajectories lead to vancomycin adaptation.
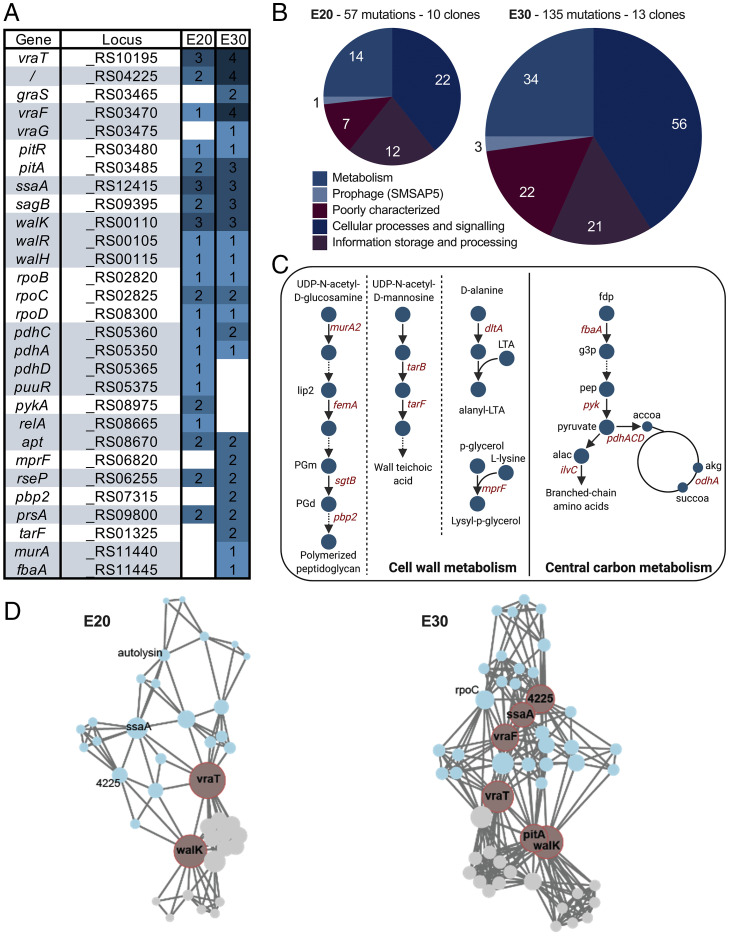
We characterized the mutational trajectory leading strain JE2 to gain vancomycin resistance by sequencing all lineages at E20 and E30. We observed increasing mutational divergence across exposure levels (SI Appendix, Fig. S1; P < 0.01). On average, two clones differ by 9.7 ± 2.7 mutated genes at E20 and by 14.2 ± 3.5 mutated genes at E30 (SI Appendix, Fig. S1). We counted a total of 109 mutations across the lineages at E30, in majority single nucleotide polymorphisms (SNPs), spanning 98 genes and 11 intergenic regions (Dataset S1 and SI Appendix, Table S2). On average, 6.5 mutations per lineage were detected at E20 and 10.9 mutations were detected at E30. In addition, we found that 18 single genes, adjacently located genes, or operons were mutated in more than one clone (Fig. 2A and SI Appendix, Table S3). At E30, most mutations in open reading frames were missense with only 13 frameshifts and five stop codons, indicating that at least some may be gain-of-function mutations (SI Appendix, Table S2). The most mutated genes corresponded to three different two-component systems—vraRST (four lineages), graXSR (two lineages), and walKR (four lineages)—and the RNA polymerase machinery (rpoB, rpoC, rpoD) (Fig. 2A). Interestingly, two adjacent genes, vraF and vraG, known to be regulated by graXSR, also featured among the most mutated loci (five lineages), along with two genes located immediately downstream of pitAR (three lineages). The second-most mutated gene in our study, SAUSA300_RS04225 (five lineages), was a putative membrane protein with an unknown function. In addition, we observed that at E30, L5 had a 54713bp deletion, resulting in the loss of the staphylococcal cassette chromosome mec (SCCmec) (Dataset S1) and that L9 had acquired a mutation in mutS causing hypermutation with the accumulation of 30 mutations between E20 and E30 compared to the average three mutations in the other clones.
Fig. 2.
Accumulated mutations across vancomycin-adapted strains at E20 and E30. (A) Top 30 most-mutated genes rank-ordered by the total number of accumulated mutations at E20 and E30 and grouped by function. (B) Mutations at E20 and E30 grouped according to COG categories. (C) Distribution of mutated genes across cell wall metabolism and central carbon metabolism pathways. lip2: lipid II, PGm: peptidoglycan monomer, PGd: peptidoglycan dimer, LTA: lipoteichoic acid, fdp: fructose-1,6-diphosphate, g3p: glyceraldehyde 3-phosphate, pep: phosphoenolpyruvate, alac: 2-acetolactate, accoa: acetyl-coenzyme A, akg: α-ketoglutarate, succoa: succinyl-coenzyme A. (D) Weighted undirected graph of mutation co-occurrence with nodes representing mutations and edges representing co-occurrence of two mutations in one lineage at E20 (Left) and E30 (Right). Communities of mutated loci with large co-occurrence frequency were computed using the Girvan–Newman method (shown in blue and gray). Node size represents harmonic centrality. Nodes in red have the largest harmonic centrality in their corresponding community.
When we grouped all mutated genes according to clusters of orthologous groups (COGs), the subsystems most affected were “cellular processes and signaling” (n = 56) and “metabolism” (n = 34) (Fig. 2B). Interestingly, genes known to be regulated by some of the mutated two-component systems encoded by walKR (n = 24), graSR (n = 17), and vraRST (n = 14) were also targeted by mutations, many of which are involved in cell wall metabolism (including pbp2, murA2, sgtB, dltA, femA, and sagB) (Fig. 2C and SI Appendix, Table S4 and Fig. S2) (15). These genes are commonly mutated in clinical VISA strains and have previously been implicated in increased vancomycin resistance (8).
Mutations related to metabolism were also prevalent in our study (n = 34). At E20, mutations accumulated around the pyruvate node including pyruvate kinase (pykA) and genes of the pyruvate dehydrogenase complex (pdhA, pdhC, and pdhD) (Fig. 2C and SI Appendix, Fig. S2), possibly leading to increased carbon flow to cell wall precursor biosynthesis (16). At E30, mutations arose in cell wall biosynthesis genes (n = 10) related to peptidoglycan biosynthesis (murA2, femA, sgtB, and pbp2), wall teichoic acid biosynthesis (tarB and tarF), alanylation of teichoic acid (dltA), and phosphatidylglycerol lysylation of the membrane (mprF) (Fig. 2C and SI Appendix, Fig. S2). Most notably, despite their common genetic background, it is evident that the lineages adapted to vancomycin through diverse mechanisms. Not only did the most frequent mutation appear in only 38% of the clones (5 out of 13), but the functional categories of genes affected by mutations also covered a large breadth of subsystems, including cell wall metabolism and transcriptional regulatory systems.
Co-occurrence networks implicate vraT and walK mutations in early adaptation and loose chronological order in mutational patterns.
To assess the chronological order and the identity of key adaptive mutations, we constructed two separate weighted undirected network graphs modeling mutation co-occurrence at E20 and E30 (Fig. 2D). Each mutation was represented as a network node, and two nodes were linked in the network if the corresponding mutations co-occurred in one strain. We proceeded to calculate the harmonic centrality (a measure of the importance of a node and its network position; see SI Appendix, Equation 1) of each mutated gene in the co-occurrence network. We found that two nodes stand out as critical early adaptive events at E20: vraT and walK (P < 0.001). Interestingly, both genes are regulators, suggesting that the first adaptive response consists of a modification of the regulatory network. At E30, additional key mutations stood out as central to the network, including locus SAUSA300_RS04225, pitA, vraF, and ssaA (centrality test, P value < 0.001). Interestingly, all four genes encode for proteins that are either localized at the membrane or secreted, suggesting that further adaptation to vancomycin involves physicochemical modifications of the membrane structure. Indeed, mutations in vraF have been linked to changes in the net surface charge (17), while mutations in pitA lead to the increased import of inorganic phosphate and increased expression of the dlt operon involved in the d-alanylation of teichoic acids (18).
Next, we asked whether adaptive strategies consisted of differences in mutational co-occurrence patterns. For this purpose, we identified node communities in both the E20 and E30 networks (SI Appendix, Table S5). We found that the communities that formed at each exposure level grouped walK, vraT, and pitA into the same community and ssaA, vraF, and SAUSA300_RS04225 into another community. The walK community consisted of 17 nodes at E20 and 25 nodes at E30 and contained all the mutations appearing in three lineages (L4, L7, and L8). Conversely, the vraF community consisted of 22 nodes at E20 and 37 nodes at E30 and contained all mutational events occurring in three different lineages (L2, L5, and L6). Thus, the network models revealed that six of the 10 clones followed two divergent evolutionary trajectories that were determined by mutational events early on in adaptation. Indeed, we noticed that the average gene level similarity within the two subcommunities was higher than the average pairwise similarity calculated for all clones (SI Appendix, Fig. S3). Finally, we also noticed that three different clones (L3, L5, and L10) converged on one path, in which a mutation in ssaA at E20 was consistently followed by a mutation in vraF at E30. Similarly, various pairs of clones converged on the following paths: vraTE20 to tarFE30 (L8, L9), walKE20 to mprFE30, and pitAE20 to mprFE30 (L4, L8). Taken together, our results suggest that there is a tendency for mutational events to follow a chronological order during vancomycin adaptation.
Vancomycin adaptation results in two opposite trajectories for β-lactam cross-susceptibility.
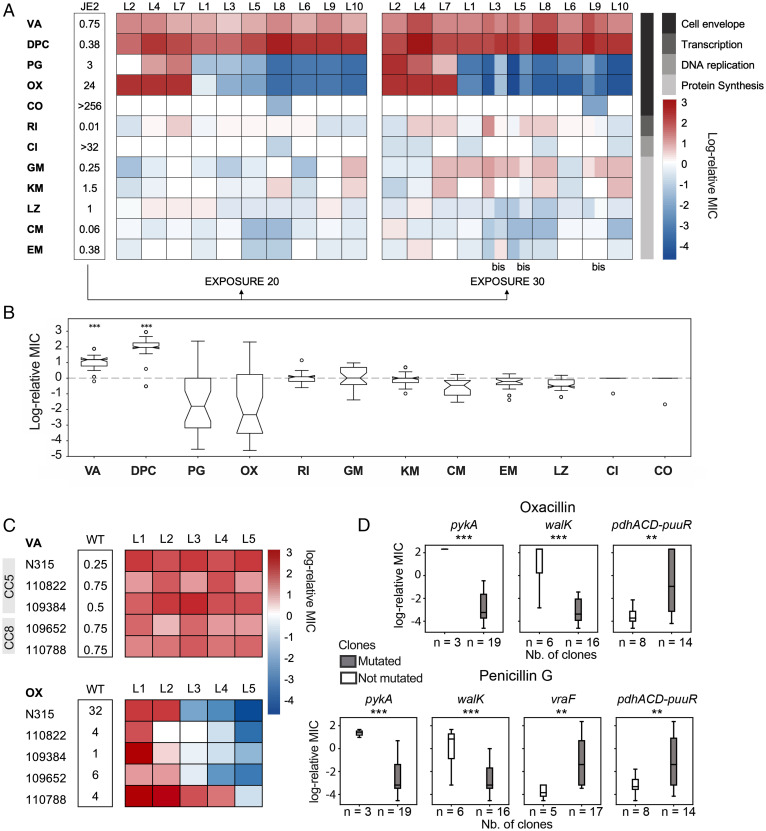
Adaptation to vancomycin is associated with decreased susceptibility to daptomycin (19, 20) and increased susceptibility to β-lactams, with the latter being termed the “seesaw effect” (21). As expected, all evolved strains had a decreased susceptibility to daptomycin. For antibiotics targeting DNA replication, protein synthesis, and transcription, we observed only few changes (Fig. 3A). Surprisingly, however, we saw two opposite penicillin susceptibility profiles (oxacillin and penicillin G) (Fig. 3 A and B). Before passage, the JE2 strain displayed an MIC toward oxacillin of 24 μg/mL but after 20 passages, oxacillin and penicillin G MICs ranged from 10 times higher in L2, L4, and L7 to 96 times lower in the other lineages. Interestingly, an initial increase or decrease in β-lactam susceptibility at E20 was accentuated at E30, suggesting that there was an early commitment in the evolution to either high or low β-lactam MIC (Fig. 3A and SI Appendix, Fig. S4). Similar divergence in adaptive response has been seen previously in both bacteria and cancer cells, where replicate lines evolved to one drug exhibit significant heterogeneity in collateral responses to second-line therapies (22–24).
Fig. 3.
JE2-derived VISA strains exhibit diverging cross-susceptibility to oxacillin and penicillin. (A) MIC of JE2-derived strains determined by Etest expressed in log-relative MIC to JE2. The bacterial target of each antibiotic is represented in gray. VA: vancomycin, DPC: daptomycin, PG: penicillin G, OX: oxacillin, CO: colistin, RI: rifampicin, CI: ciprofloxacin, GM: gentamicin, KM: kanamycin, LZ: linezolid, CM: clindamycin, EM: erythromycin. “bis” refers to additional clones L3_E30bis, L5_E30bis and L9_E30bis. (B) Box plot representing the variation in log-relative MIC across JE2-derived VISA strains. Strains showed significantly increased MIC levels for vancomycin and daptomycin with respect to wild type (Student’s t test, P < 0.001) but diverging susceptibility to penicillin and oxacillin. (C) Vancomycin and oxacillin MIC of clinical isolates N315, 110822, 109384, 109652, and 110788 belonging to clonal complexes CC5 and CC8 passaged on increasing concentrations of vancomycin for 20 exposure cycles in five replicate lineages each. The MIC is expressed in log-relative MIC to the corresponding ancestor strain. (D) Point biserial test to identified correlations between antibiotic susceptibility (log-relative MIC levels with respect to wild-type strain JE2 measured by Etest) and presence or absence of mutations.
In a recent study it was elegantly shown that the genetic background of strains impacts the trajectory of vancomycin adaptation, with CC5 strains adapting faster than CC8 (6). To examine whether divergence in β-lactam susceptibility upon vancomycin adaptation is limited to strain JE2, we adapted five clinical VSSA strains belonging to the clonal complex CC8 (the strain to which strain JE2 belongs) and CC5, both of which are predominant VISA clonal complexes (8). With 20 exposure cycles, we adapted each strain to vancomycin in five independent parallel evolutions and observed that all strains exhibited increased vancomycin resistance across lineages while individual lineages showed increased, decreased, or unchanged oxacillin resistance profiles (Fig. 3C). Thus, our observations from JE2 also apply to clinical strains belonging to various clonal complexes.
We next asked whether mutational patterns across the JE2 lineages could explain β-lactam susceptibility. For this purpose, we performed a multiway ANOVA (Fig. 3D) and found that mutations in pykA and walK had the biggest effect on measured susceptibility to oxacillin and penicillin G (P ∼= 0.0001 and P ∼= 0.0002, respectively), followed by mutations in vraF (P ∼= 0.003) and the pyruvate dehydrogenase complex pdhACD (P = 0.001, 0.002, respectively). However, while the presence of mutations in pykA and walK correlated with increased resistance, mutations in either vraF or pdhACD correlated with increased susceptibility to both β-lactams. Indeed, only clones L4 and L7 carried a deletion in the pykA gene and a single nucleotide variant in walK, while clones L3, L5, L9, and L10 accumulated various mutations in vraF and clones L6, L8, and L10 accumulated mutations in pdhACD. Taken together, our results suggest that separate mechanisms are at play in the progression of cross-susceptibility versus cross-resistance and that collateral sensitivity to β-lactams is only one of three possible outcomes during vancomycin adaptation.
Vancomycin adaptation induces diverse transcriptional reprogramming that cannot be readily linked to mutational patterns.
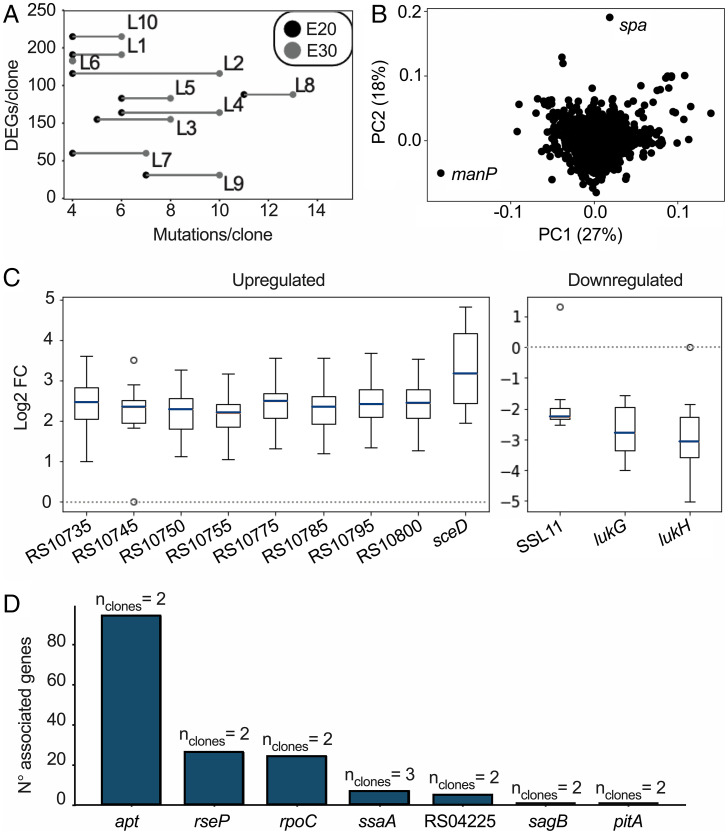
We next addressed whether there was a transcriptional response associated with vancomycin adaptation. To this end, we extracted the expression profiles of all 10 clones at E20. On average, 134 ± 55 genes were differentially expressed per evolved clone with respect to strain JE2, ranging from 31 genes in L9 to 215 genes in L10 (Fig. 4A). Notably, expression patterns were highly divergent across strains and a principal component analysis failed to cluster the transcriptional profiles across strains (Fig. 4B). Indeed, while 598 genes showed significant differential expression (DEGs; > 1.5 log2 fold change) in at least one clone, only sceD and lukG were significantly up-regulated and down-regulated (respectively) across the board (Fig. 4C). In 9 out of 10 clones, lukH and ssl11 were down-regulated while eight colocated φSa3 prophage genes were up-regulated. SceD is a lytic transglycosylase involved in cell wall turnover, growth, and cell separation (25), while LukGH (a bicomponent leukotoxin [26]), and the staphylococcal superantigen-like protein 11 (SSL11) are virulence factors with cytotoxic effects against immune cells (27). Prophage Sa3 (φSa3) is a prophage that has previously been found to excise during vancomycin adaptation (7).
Fig. 4.
Summary of the RNA sequencing analyses. (A) Scatterplot showing the number of mutations in E20 and in E30 versus the number of DEGs in E20. (B) Principal component (PC) analysis of the genome-wide transcriptional profile. Each gene is represented in this plot by one dot and is processed as a 10-dimensional vector of log2 fold change values of normalized counts against the expression profile of strain JE2. (C) Subset of genes showing a log2 fold change (FC) of more than 1.5 in at least nine out of 10 clones. (D) Association between gene expression and mutations; e.g., 94 genes have a common transcriptional response when comparing two clones with an apt mutation.
We proceeded to search for links between genetic and transcriptional changes (Fig. 4D). For each mutated gene, we separated clones into two groups according to whether they carried a mutation in that gene at E20. We compared the expression profiles for all genes between the two groups and extracted the genes showing significant differential expression between the two groups (> 1.5 log2 fold change, P < 0.05, alpha = 0.05). To our surprise, we observed very few associations between mutational events and differential expression profiles. In fact, it was only possible to observe correlating transcriptional responses for six genes mutated in two or three clones (Fig. 4D), including ssaA, a cell wall amidase, and rpoC. Thus, identical mutations led to different transcriptional responses and even though adaptation to vancomycin has repeatedly been associated with mutations in transcriptional regulators, we could not find a common transcriptional pattern explaining these mutations. Taken together, our results demonstrate that vancomycin adaptation at the transcriptional level is complex, multifactorial, and divergent.
Transcriptional decomposition reveals differential gene expression in vancomycin-adapted clones with low penicillin susceptibility.
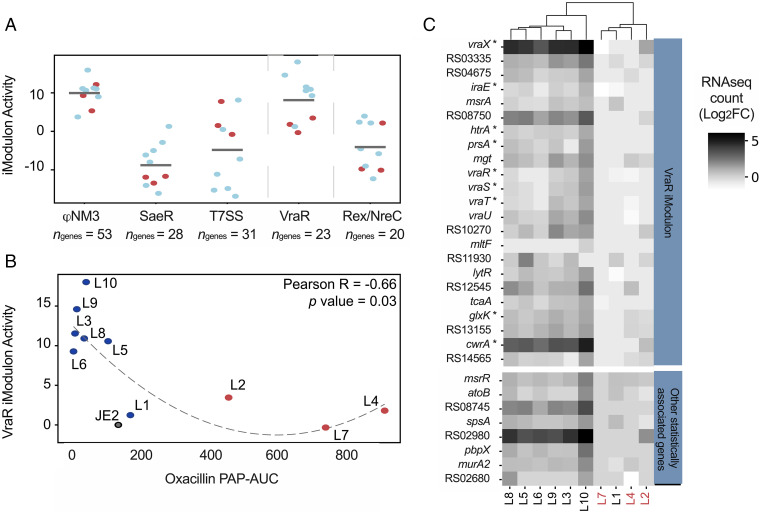
We next asked whether vancomycin adaptation had a nonlinear effect on expression. For this purpose, we turned to ICA, by which transcriptional regulatory networks can be “decomposed” into iModulons, or coregulated, independently modulated sets of genes (28, 29). Analysis of the transcriptional regulatory network of the vancomycin adapted clones at E20 revealed that the five most impacted iModulons in adapted strains were φSa3, SaeR (Two-component system (TCS) regulating > 20 virulence genes), T7SS (Type 7 secretion system), VraR (TCS linked to vancomycin resistance), and Rex/NreC (TCS controlling nitrate and nitrite reductase operons [30]) (Fig. 5A and Dataset S2). Interestingly, the first three iModulons were involved in virulence, with the SaeR iModulon containing both sceD and lukGH. Our findings corroborate previous findings demonstrating an altered virulence profile of VISA strains (31).
Fig. 5.
Transcriptional profiles related to vancomycin adaptation and β-lactam cross-sensitivity in JE2. (A) ICA of the mutants’ transcriptional profiles reveal five iModulons (independently modulated sets of genes) with the largest changes in activity with respect to wild type. The scatterplot shows the activity levels of each iModulon in each clone. Blue and red dots represent clones with cross-susceptibility and cross-resistance to oxacillin, respectively. (B) The VraR iModulon is the only iModulon showing significant correlation between activity levels in clones and corresponding susceptibility to oxacillin. A decrease in VraR iModulon activity explains increased susceptibility to oxacillin. (C) Cluster map of the expression levels (log2 fold change [FC] with respect to wild type) in the VraR iModulon genes. Genes showing correlated expression levels with MIC are annotated with a star. Additional genes not forming part of the VraR iModulon but that nonetheless also reveal significant correlation with MIC levels are shown separately in the bottom heatmap. Clones are color coded according to cross-resistance to oxacillin (red = lineages with high-oxacillin MIC).
Another question was whether transcriptional patterns were linked with oxacillin susceptibility. Initial analysis of gene expression profiles showed that the expression of mecA encoding the alternative penicillin binding protein, PBP2a, did not explain the antibiotic susceptibility differences between clones (Dataset S3). To identify whether changes in iModulon activity were reflected in MIC variations, we systematically searched for correlations between iModulon activity levels and log-fold MIC values. Our search returned a single inverse correlation between the VraR iModulon and oxacillin susceptibility (Pearson R = −0.66, P = 0.03) (Fig. 5B). Notably, while increased activity of the VraR iModulon was associated with decreased susceptibility to oxacillin, the reverse was not true. Thus, VraR iModulon activity only explained increased susceptibility and not increased resistance to oxacillin.
The VraR iModulon contains 23 genes (Fig. 5C), 18 of which have been previously annotated as being part of the vraR regulon (9). Of the 23 genes, we found 18 genes for which the expression level showed significant correlation with oxacillin MIC (Spearman R > 0.7; Dataset S4), of which 5 were mutated in at least one clone: mgt, SAUSA300_RS08750, prsA, htrA, and vraT (SI Appendix, Fig. S5). Notably the vraR genes were not mutated in any of our evolved strains. Instead, VraT, which is responsible for the activation of the VraSR regulon (9), was mutated in three lineages with a low-oxacillin MIC at E20 but was also mutated at E30 in the high-oxacillin MIC clone L7. To examine the individual contributions of the vraT mutations and the overexpression of VraR-regulated genes, we reconstructed the vraT mutations in JE2 and overproduced selected VraR regulated genes as well as SceD that we had found to be overexpressed in all lineages (SI Appendix, Tables S6 and S7). Mutations in vraT resulted in increased vancomycin and oxacillin MIC, regardless of whether they originated from a lineage with high- or low-oxacillin MIC (SI Appendix, Table S6). Overexpression of selected VraR-regulated genes did not individually impact susceptibility to antibiotics, while VraR overexpression, as reported in the literature, increased oxacillin MIC (32). We previously showed that clones carrying mutations in walK displayed increased oxacillin MIC (Fig. 3D). The introduction of these mutations in JE2 resulted in increased vancomycin MIC and, surprisingly, decreased oxacillin MIC (SI Appendix, Table S6). These findings show that mutations in other genes affect the expression and activity of vraR and walK and that the susceptibility phenotypes observed for the adapted lineages depend on epistasis, where the effect of one mutation depends on the presence of other mutations.
Correlations between susceptibility to penicillin and cell wall inhibitors.
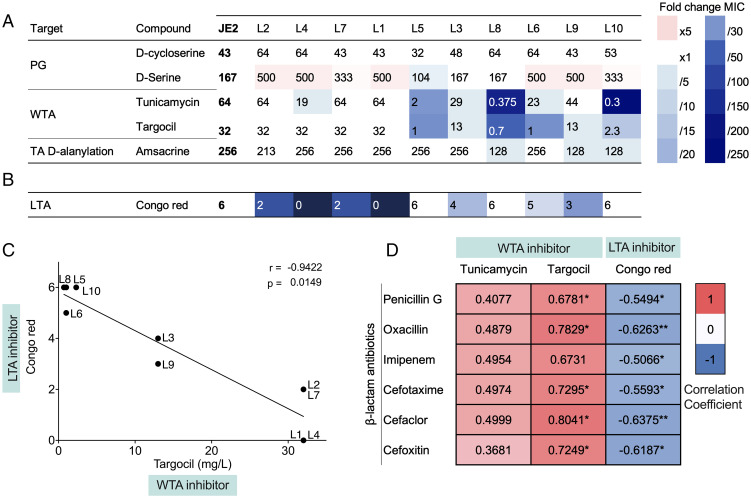
Throughout our investigations of vancomycin-adapted lineages, results had pointed to genes suspected to cause significant changes in the cell envelope as being important (Figs. 2, 3, and 5). Therefore, we finally examined susceptibility to cell wall inhibitors targeting either the peptidoglycan or the teichoic acids that decorate the cell wall and are divided into lipoteichoic (LTA) and wall teichoic acids (WTA). While all lineages showed equal susceptibility to peptidoglycan synthesis inhibitors, there were striking differences toward inhibitors of LTA and WTA (Fig. 6 A and B). In fact, there was an inverse correlation between susceptibility to LTA and WTA inhibitors across lineages (Fig. 6C). Interestingly, lineages with high β-lactam MIC (L2, L4, and L7) were more susceptible than JE2 and the remaining lineages to congo red that inhibits the LTA synthase, LtaS (33), while the lineages showing low β-lactam MIC were more susceptible to the WTA inhibitors targocil and tunicamycin affecting WTA biogenesis (34) (Fig. 6D). This finding is interesting because in MRSA strains, inhibitors of WTA increase susceptibility to β-lactam antibiotics (35) and in the absence of WTA, synthetic lethality is observed for mutants also lacking LTA (36). Thus, the increase or decrease in β-lactam susceptibility of vancomycin-adapted lineages may be related to teichoic acid structures.
Fig. 6.
Susceptibility of JE2-derived VISA strains to cell envelope–targeting compounds. (A) Susceptibility to compounds targeting the peptidoglycan (PG), WTA, teichoic acid D-alanylation, and LTA. Susceptibility to D-cycloserine, tunicamycin, targocil, and amsacrine is expressed in mg/L. D-serine susceptibility is expressed in mM. For congo red susceptibility testing, serial dilutions of overnight cultures were spotted on TSA plates supplemented with 0.1% congo red. The results show the highest bacterial dilution at which growth was observed, expressed in 10-#. Gradients of red and blue highlight fold change increases and decreases in MIC compared to parental strain JE2. (B) Linear regression showing the correlation between targocil, tunicamycin, and congo red susceptibility. MIC is expressed in mg/L for targocil and tunicamycin. Susceptibility is expressed as the highest dilution at which growth was observed in 10-# for congo red. r: Pearson correlation coefficient, p: P value. (C) Pearson correlation matrix showing correlation coefficient for pairs of beta-lactam antibiotics penicillin G, oxacillin, imipenem, cefotaxime, cefaclor, and imipenem, and WTA/LTA inhibitors tunicamycin, targocil, and congo red. Pearson correlation coefficient r = −1, r = 0, r = 1. P < 0.1*, P < 0.01**.
Discussion
Antibiotic resistance is commonly associated with the acquisition of well-defined resistance genes or with specific point mutations affecting central microbial processes, but for some antibiotics, reduced susceptibility requires multiple mutations and the genes affected can vary considerably. One example is colistin resistance in Pseudomonas aeruginosa that results from multiple mutations impacting, among others, transcriptional regulators that function as nodes in potentiating further evolution (37). Another example is decreased vancomycin susceptibility in the S. aureus VISA strains (8). Here we have examined the mutational and transcriptional landscape of S. aureus VISA by evolving 10 lineages in parallel under the same selection pressure starting from a single VSSA strain (JE2). We found the initial response to be dominated by mutations of transcriptional regulators across lineages while the secondary response was characterized by mutations of the cell wall synthesis apparatus in genes such as pbp2, prsA, sgtB, mprF, and dltA. Notably, our study recapitulates known adaptive mutations that have appeared in clinical VISA isolates (e.g., mutations in vraT, graS, vraFG, walKR, and rpoBCD) as well as unreported adaptive mutations. Furthermore, it corroborates a recent study where massive genome sequencing of vancomycin-adaptive lineages pointed to mutations in walK, rpoC, vraS, and yvqF/vraT as being statistically associated with VISA in a CC5 strain although only walK mutations were significant in a CC8 strain (6).
The transcriptional responses of our 10 adapted lineages turned out to be highly lineage-specific, with only two genes showing differential expression across lineages compared to JE2: sceD (up-regulated) and lukG (down-regulated). LukGH is a bicomponent leukotoxin previously associated with VISA (26), and SceD is a lytic transglycosylase that is often overexpressed in VISA and hVISA strains (14, 38–40). We further investigated potential associations between mutations and transcriptional patterns. Again, the analysis revealed a surprisingly low number of links between genomic and transcriptional alterations (Fig. 4D). Thus, different mutations may have nonlinear downstream transcriptional effects, and mutations in different genes provide similar transcriptional changes.
To dissect the nonlinear transcriptional contributions to the VISA phenotype, we used ICA. Here, our analysis pointed to virulence as being the most perturbed functional group, with three independent virulence systems affected including φSa3, SaeR, and T7SS. SaeR is the response regulator of the SaeRS two-component system that regulates more than 20 virulence genes in S. aureus (41). The expression of SaeR is induced by vancomycin (42), but only a few studies have linked it with VISA (43, 44). Notably, the transcriptional changes we observed for sceD and lukGH match with low SaeR activity, as sceD expression is greatly induced in a saeR mutant (25) and expression of the lukGH toxin genes is positively regulated by the system (45). We also found that our vancomycin-evolved lineages exhibited increased activity of the VraR iModulon compared to JE2. Increased expression of the VraSR regulon is associated with VISA strains (8, 46, 47) and conversely, deletion of the vraSR operon enhances sensitivity to vancomycin and other cell wall–targeting antibiotics (9, 48). Also revealed by the iModulon analysis was NreC/Rex, which controls the nitrate reductase and nitrite reductase operons (30) and recently was demonstrated to impact oxacillin resistance in MRSA strains (49).
Previous studies have shown that when MRSA strains become VISA, they tend to lose β-lactam resistance, and it has been suggested that this collateral sensitivity may enable the clinical use of β-lactams when treating VISA infections (21). We find that S. aureus employs multiple evolutionary trajectories when adapting to vancomycin, resulting in strains with either significantly increased or decreased MIC toward oxacillin and penicillin. The diverging β-lactam susceptibility was not explained by altered expression of pbp2 or mecA encoding PBP2a (Dataset S3). but increased VraR iModulon activity correlated with low β-lactam MIC (Fig. 5B) while mutations in walK correlated with high β-lactam MIC (Fig. 3D). However, neither the overexpression of VraR- or VraR-regulated genes nor the re-creation of vraT or walK mutations recapitulated the resistance patterns of the adapted strains, highlighting that epistasis is central in VISA strains.
Since the VraSR/VraT and WalRK two-component systems respond to cell wall stress (11, 50) we also examined the susceptibility to cell envelope inhibitors and found that lineages with high β-lactam MIC showed differential susceptibility to an LTA inhibitor, whereas the lineages with low MIC were sensitive to WTA inhibitors. This finding was interesting because MRSA strains become susceptible to β-lactam antibiotics in the presence of WTA inhibitors (35) and in the absence of WTA, synthetic lethality is observed for mutants also lacking LTA, VraT, VraFG, and GraRS (36). Our results could indicate that WTA and LTA are differentially affected during vancomycin adaptation with an impact on β-lactam susceptibility. If so, such differences may also impact virulence because teichoic acids are central in defensin resistance and for recognition by the host immune system (51).
Collectively, our work describes an impressive outcome space for a single MRSA strain when evolving to VISA, including a divergent effect on susceptibility to β-lactam antibiotics. Collateral drug sensitivity has been observed for both bacteria and cancer cells (22, 24). However, similar to our findings, a recent study suggested that collateral sensitivity is not universal (23). It has also been noted that the path leading to collateral sensitivity may not be repeatable (24) and in therapy, optimal drug use may be more efficacious than cycles of collateral drug sensitivity in maintaining lower long-term drug resistance (22). We show here that by applying ICA in dissecting contributions from mutations and multiple transcriptional regulators we can dissect the underlying physiology of collateral sensitivity and identify overlooked pathways as being central contributors to drug resistance. Our results provide a foundation for future studies into how resistance to cell wall antibiotics evolves and identify targets that may inspire combination therapies.
Materials and Methods
Bacterial strains and growth conditions.
The bacterial strains used in this study are listed in SI Appendix, Table S8. The construction of strains is detailed in the SI Appendix. Tryptic soy agar (TSA) and tryptic soy broth (TSB) were used for routine growth of bacterial strains. Brain–heart infusion (BHI) agar, Mueller-Hinton (MH) agar, and MH broth were used for PAP and antibiotic susceptibility testing. Sheep blood agar was used for hemolysis assays. The strains were incubated at 37 °C with shaking unless otherwise specified. Antibiotic stock solutions were prepared following the recommendations of the manufacturer.
Serial transfer experiments.
The MRSA strain JE2, a plasmid-cured derivative of USA300 LAC, was grown on increasing concentrations of vancomycin in TSB medium in 10 independent parallel lineages (L1 to L10) using a two-step protocol derived from Rodriguez de Evgrafov et al. (52) (SI Appendix, Fig. S6). This process was repeated for 30 exposure cycles (E), and thorough characterization was conducted every 10 cycles at timepoints E10, E20, and E30. SI Appendix contains the detailed protocol.
Susceptibility testing.
MICs for various antibiotics were obtained by Etest (Biomérieux) or broth microdilution. Broth microdilution MIC testing was performed following Clinical and Laboratory Standards Institute guidelines (53). Etests were used according to the instructions of the manufacturer with S. aureus strain ATCC 29213 as the control.
PAPs.
Population analysis was performed as described previously (13). Overnight cultures were serial diluted to 10−6 and dilutions were plated on BHI agar containing a series of concentrations of vancomycin or oxacillin. Bacterial colonies were counted after incubation of the plates at 37 °C for 48 h. The AUC was calculated using GraphPad Prism 7. For vancomycin population analysis, the AUC of the tested strain was divided by the AUC of Mu3, a standard hVISA strain. This ratio was used to determine the status of the strain: An AUC ratio to mu3 of 0.9 and above characterizes a hVISA strain, while an AUC ratio superior or equal to 1.3 detects a VISA strain (13). Population analysis was performed in 3 to 5 biological replicates.
Generation times.
Overnight cultures were diluted to 5 × 105 CFU/mL in TSB and incubated at 37 °C in a Bioscreen C microbiological reader (LabSystems) for 24 h with medium shaking. Optical density (OD) measurements at 600 nm were automatically recorded every 10 min. Each strain was tested in biological triplicates and technical quadruplicates. Generation times were calculated using the program GrowthRates 3.0 developed by Hall et al. (54).
Hemolysis assays.
Overnight cultures were diluted, spotted on TSA supplemented with 5% sheep blood, and incubated for 24 h at 37 °C.
Genome sequencing and mutation analysis.
DNA sequencing was performed on the JE2 wild-type strain and the JE2-derived vancomycin-adapted clones. Reads were mapped against the S. aureus USA300_FPR3757 reference genome (GenBank accession no. NC_07793). The variants identified during alignment of JE2 WT to the reference genome were excluded in the variant call for vancomycin-adapted clones. Further details can be found in SI Appendix.
RNA sequencing and gene expression analysis.
Strains were grown in triplicate in TSB and harvested at OD 600 nm = 0.5. RNA was extracted using the RNeasy Mini Kit (Qiagen). After ribosomal RNA depletion, 250 to 300 bp insert strand-specific cDNA libraries were prepared and sequenced using Illumina NovaSeq platforms with a paired-end 150 bp sequencing strategy. The transcriptome analysis was performed using FastQC for quality control, Bowtie2 for mapping, htseq-count to count the number of mapped reads to each gene, and DeSeq2 to assess the differential expression between ancestor and evolved strains.
Functional annotation of mutated loci.
We used Eggnog-mapper to determine the functional distribution of mutated loci (55). Metabolic genes affected by mutations were further annotated with subsystems and catalytic activity using a curated genome-scale model of metabolism (56). Functional loci affected by mutations were also mapped to transcriptional regulons of two-component systems pulled from a variety of literature sources; graSR (15), lytSR (57), nreABC (30), saeRS (45), arlRS (45, 58), walKR (59), and vraRST (9).
Mutation co-occurrence analysis.
To assess the co-occurrence of mutations across strains, we built a weighted graph in Networkx in which mutations were represented as nodes and the co-occurrence of two mutations in strains were represented by edges. To inspect the chronology of mutation emergence, we constructed two separate weighted undirected graphs for each exposure level. Details on the analysis can be found in SI Appendix.
ICA of transcriptomic profile.
RNA sequencing and the ICA pipeline used to calculate the iModulon structure and activities have been described in full detail in previous publications (12, 29). Here, normalized transcripts per million from 33 new RNA sequencing profiles were calculated concatenated to previously published 108 profiles (29). The analysis is detailed in SI Appendix.
Analysis of statistical associations between mutations and phenotypes.
We screened all the mutations that appeared in two or more clones against measured minimal inhibitory concentrations using the point biserial test. Mutations that scored a P value below 0.05 were then tested using a multiway ANOVA.
Supplementary Material
Acknowledgments
This project has received funding from the European Union’s Horizon 2020 research, number 765147; the National Institutes of Allergies and Infectious Disease, grant number AI124316; and the Olav Thon foundation. We thank Statens Serum Institut for providing us with MRSA clinical isolates.
Footnotes
The authors declare no competing interest.
This article is a PNAS Direct Submission.
This article contains supporting information online at https://www.pnas.org/lookup/suppl/doi:10.1073/pnas.2118262119/-/DCSupplemental.
Data Availability
Sequence data have been deposited in the European Nucleotide Archive under study accession number PRJEB52876 (60) and are publicly available. All other study data are included in the article and supporting information.
References
- 1.McGuinness W. A., Malachowa N., DeLeo F. R., Vancomycin resistance in Staphylococcus aureus. Yale J. Biol. Med. 90, 269–281 (2017). [PMC free article] [PubMed] [Google Scholar]
- 2.Roch M., et al. , Exposure of Staphylococcus aureus to subinhibitory concentrations of β-lactam antibiotics induces heterogeneous vancomycin-intermediate Staphylococcus aureus. Antimicrob. Agents Chemother. 58, 5306–5314 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Howden B. P., Davies J. K., Johnson P. D. R., Stinear T. P., Grayson M. L., Reduced vancomycin susceptibility in Staphylococcus aureus, including vancomycin-intermediate and heterogeneous vancomycin-intermediate strains: Resistance mechanisms, laboratory detection, and clinical implications. Clin. Microbiol. Rev. 23, 99–139 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Howden B. P., Johnson P. D. R., Ward P. B., Stinear T. P., Davies J. K., Isolates with low-level vancomycin resistance associated with persistent methicillin-resistant Staphylococcus aureus bacteremia. Antimicrob. Agents Chemother. 50, 3039–3047 (2006). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Mwangi M. M., et al. , Tracking the in vivo evolution of multidrug resistance in Staphylococcus aureus by whole-genome sequencing. Proc. Natl. Acad. Sci. U.S.A. 104, 9451–9456 (2007). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Su M., et al. , Effect of genetic background on the evolution of vancomycin-intermediate Staphylococcus aureus (VISA). PeerJ 9, e11764 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Machado H., et al. , Environmental conditions dictate differential evolution of vancomycin resistance in Staphylococcus aureus. Commun. Biol. 4, 793 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Hu Q., Peng H., Rao X., Molecular events for promotion of vancomycin resistance in vancomycin intermediate Staphylococcus aureus. Front. Microbiol. 7, 1601 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Boyle-Vavra S., Yin S., Jo D. S., Montgomery C. P., Daum R. S., VraT/YvqF is required for methicillin resistance and activation of the VraSR regulon in Staphylococcus aureus. Antimicrob. Agents Chemother. 57, 83–95 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Falord M., Karimova G., Hiron A., Msadek T., GraXSR proteins interact with the VraFG ABC transporter to form a five-component system required for cationic antimicrobial peptide sensing and resistance in Staphylococcus aureus. Antimicrob. Agents Chemother. 56, 1047–1058 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Dubrac S., Boneca I. G., Poupel O., Msadek T., New insights into the WalK/WalR (YycG/YycF) essential signal transduction pathway reveal a major role in controlling cell wall metabolism and biofilm formation in Staphylococcus aureus. J. Bacteriol. 189, 8257–8269 (2007). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Sastry A. V., et al. , The Escherichia coli transcriptome mostly consists of independently regulated modules. Nat. Commun. 10, 5536 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Wootton M., et al. , A modified population analysis profile (PAP) method to detect hetero-resistance to vancomycin in Staphylococcus aureus in a UK hospital. J. Antimicrob. Chemother. 47, 399–403 (2001). [DOI] [PubMed] [Google Scholar]
- 14.Cafiso V., et al. , Modulating activity of vancomycin and daptomycin on the expression of autolysis cell-wall turnover and membrane charge genes in hVISA and VISA strains. PLoS One 7, e29573 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Herbert S., et al. , Molecular basis of resistance to muramidase and cationic antimicrobial peptide activity of lysozyme in Staphylococci. PLoS Pathog. 3, e102 (2007). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Gardner S. G., Marshall D. D., Daum R. S., Powers R., Somerville G. A., Metabolic mitigation of Staphylococcus aureus vancomycin intermediate-level susceptibility. Antimicrob. Agents Chemother. 62, e01608-17 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Meehl M., Herbert S., Götz F., Cheung A., Interaction of the GraRS two-component system with the VraFG ABC transporter to support vancomycin-intermediate resistance in Staphylococcus aureus. Antimicrob. Agents Chemother. 51, 2679–2689 (2007). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Mechler L., et al. , Daptomycin tolerance in the Staphylococcus aureus pitA6 mutant is due to upregulation of the dlt operon. Antimicrob. Agents Chemother. 60, 2684–2691 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Cui L., Tominaga E., Neoh H. M., Hiramatsu K., Correlation between reduced daptomycin susceptibility and vancomycin resistance in vancomycin-intermediate Staphylococcus aureus. Antimicrob. Agents Chemother. 50, 1079–1082 (2006). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Patel J. B., Jevitt L. A., Hageman J., McDonald L. C., Tenover F. C., An association between reduced susceptibility to daptomycin and reduced susceptibility to vancomycin in Staphylococcus aureus. Clin. Infect. Dis. 42, 1652–1653 (2006). [DOI] [PubMed] [Google Scholar]
- 21.Ortwine J. K., Werth B. J., Sakoulas G., Rybak M. J., Reduced glycopeptide and lipopeptide susceptibility in Staphylococcus aureus and the “seesaw effect”: Taking advantage of the back door left open? Drug Resist. Updat. 16, 73–79 (2013). [DOI] [PubMed] [Google Scholar]
- 22.Maltas J., Wood K. B., Pervasive and diverse collateral sensitivity profiles inform optimal strategies to limit antibiotic resistance. PLoS Biol. 17, e3000515 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Nichol D., et al. , Antibiotic collateral sensitivity is contingent on the repeatability of evolution. Nat. Commun. 10, 334 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Scarborough J. A., et al. , Identifying states of collateral sensitivity during the evolution of therapeutic resistance in Ewing’s sarcoma. iScience 23, 101293 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Stapleton M. R., et al. , Characterization of IsaA and SceD, two putative lytic transglycosylases of Staphylococcus aureus. J. Bacteriol. 189, 7316–7325 (2007). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Yanai M., et al. , Separately or combined, LukG/LukH is functionally unique compared to other staphylococcal bicomponent leukotoxins. PLoS One 9, e89308 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Chen C., Yang C., Barbieri J. T., Staphylococcal superantigen-like protein 11 mediates neutrophil adhesion and motility arrest, a unique bacterial toxin action. Sci. Rep. 9, 4211 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Rychel K., Sastry A. V., Palsson B. O., Machine learning uncovers independently regulated modules in the Bacillus subtilis transcriptome. Nat. Commun. 11, 6338 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Poudel S., et al. , Revealing 29 sets of independently modulated genes in Staphylococcus aureus, their regulators, and role in key physiological response. Proc. Natl. Acad. Sci. U.S.A. 117, 17228–17239 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Schlag S., et al. , Characterization of the oxygen-responsive NreABC regulon of Staphylococcus aureus. J. Bacteriol. 190, 7847–7858 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Jin Y., et al. , Comparative analysis of virulence and toxin expression of vancomycin-intermediate and vancomycin-sensitive Staphylococcus aureus strains. Front. Microbiol. 11, 596942 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Boyle-Vavra S., Yin S., Daum R. S., The VraS/VraR two-component regulatory system required for oxacillin resistance in community-acquired methicillin-resistant Staphylococcus aureus. FEMS Microbiol. Lett. 262, 163–171 (2006). [DOI] [PubMed] [Google Scholar]
- 33.Vickery C. R., Wood B. M., Morris H. G., Losick R., Walker S., Reconstitution of S. aureus lipoteichoic acid synthase activity identifies Congo red as a selective inhibitor. J. Am. Chem. Soc. 140, 876–879 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Rajagopal M., Walker S., Envelope structures of gram-positive bacteria. Curr. Top. Microbiol. Immunol. 404, 1–44 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Campbell J., et al. , Synthetic lethal compound combinations reveal a fundamental connection between wall teichoic acid and peptidoglycan biosyntheses in Staphylococcus aureus. ACS Chem. Biol. 6, 106–116 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Santa Maria J. P. Jr., et al. , Compound-gene interaction mapping reveals distinct roles for Staphylococcus aureus teichoic acids. Proc. Natl. Acad. Sci. U.S.A. 111, 12510–12515 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Jochumsen N., et al. , The evolution of antimicrobial peptide resistance in Pseudomonas aeruginosa is shaped by strong epistatic interactions. Nat. Commun. 7, 13002 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Drummelsmith J., Winstall E., Bergeron M. G., Poirier G. G., Ouellette M., Comparative proteomics analyses reveal a potential biomarker for the detection of vancomycin-intermediate Staphylococcus aureus strains. J. Proteome Res. 6, 4690–4702 (2007). [DOI] [PubMed] [Google Scholar]
- 39.Kuroda M., et al. , IS256-mediated overexpression of the WalKR two-component system regulon contributes to reduced vancomycin susceptibility in a Staphylococcus aureus clinical isolate. Front. Microbiol. 10, 1882 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Pieper R., et al. , Comparative proteomic analysis of Staphylococcus aureus strains with differences in resistance to the cell wall-targeting antibiotic vancomycin. Proteomics 6, 4246–4258 (2006). [DOI] [PubMed] [Google Scholar]
- 41.Liu Q., Yeo W.-S., Bae T., The SaeRS two-component system of Staphylococcus aureus. Genes (Basel) 7, 81 (2016). [Google Scholar]
- 42.Hessling B., et al. , Global proteome analysis of vancomycin stress in Staphylococcus aureus. Int. J. Med. Microbiol. 303, 624–634 (2013). [DOI] [PubMed] [Google Scholar]
- 43.Dai Y., et al. , VraR binding to the promoter region of agr inhibits its function in vancomycin-intermediate Staphylococcus aureus (VISA) and heterogeneous VISA. Antimicrob. Agents Chemother. 61, e02740-16 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Park C., et al. , Genomic analysis of heterogeneous vancomycin-intermediate Staphylococcus aureus strains from different clonal lineages in South Korea. Microb. Drug Resist. 27, 1271–1281 (2021). [DOI] [PubMed] [Google Scholar]
- 45.Rogasch K., et al. , Influence of the two-component system SaeRS on global gene expression in two different Staphylococcus aureus strains. J. Bacteriol. 188, 7742–7758 (2006). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Gardete S., et al. , Genetic pathway in acquisition and loss of vancomycin resistance in a methicillin resistant Staphylococcus aureus (MRSA) strain of clonal type USA300. PLoS Pathog. 8, e1002505 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.McAleese F., et al. , Overexpression of genes of the cell wall stimulon in clinical isolates of Staphylococcus aureus exhibiting vancomycin-intermediate S. aureus-type resistance to vancomycin. J. Bacteriol. 188, 1120–1133 (2006). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.McCallum N., Meier P. S., Heusser R., Berger-Bächi B., Mutational analyses of open reading frames within the vraSR operon and their roles in the cell wall stress response of Staphylococcus aureus. Antimicrob. Agents Chemother. 55, 1391–1402 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Panchal V. V., et al. , Evolving MRSA: High-level β-lactam resistance in Staphylococcus aureus is associated with RNA polymerase alterations and fine tuning of gene expression. PLoS Pathog. 16, e1008672 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Kuroda M., et al. , Two-component system VraSR positively modulates the regulation of cell-wall biosynthesis pathway in Staphylococcus aureus. Mol. Microbiol. 49, 807–821 (2003). [DOI] [PubMed] [Google Scholar]
- 51.Fedtke I., Götz F., Peschel A., Bacterial evasion of innate host defenses—The Staphylococcus aureus lesson. Int. J. Med. Microbiol. 294, 189–194 (2004). [DOI] [PubMed] [Google Scholar]
- 52.Rodriguez de Evgrafov M., Gumpert H., Munck C., Thomsen T. T., Sommer M. O. A., Collateral resistance and sensitivity modulate evolution of high-level resistance to drug combination treatment in Staphylococcus aureus. Mol. Biol. Evol. 32, 1175–1185 (2015). [DOI] [PubMed] [Google Scholar]
- 53.Clinical and Laboratory Standards Institute, Performance standards for antimicrobial susceptibility testing, ed. 32, document M100-S20. https://clsi.org/standards/products/microbiology/documents/m100/. Accessed 27 June 2022. [Google Scholar]
- 54.Hall B. G., Acar H., Nandipati A., Barlow M., Growth rates made easy. Mol. Biol. Evol. 31, 232–238 (2014). [DOI] [PubMed] [Google Scholar]
- 55.Huerta-Cepas J., et al. , Fast genome-wide functional annotation through orthology assignment by eggNOG-mapper. Mol. Biol. Evol. 34, 2115–2122 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Seif Y., et al. , A computational knowledge-base elucidates the response of Staphylococcus aureus to different media types. PLOS Comput. Biol. 15, e1006644 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Sharma-Kuinkel B. K., et al. , The Staphylococcus aureus LytSR two-component regulatory system affects biofilm formation. J. Bacteriol. 191, 4767–4775 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Liang X., et al. , Global regulation of gene expression by ArlRS, a two-component signal transduction regulatory system of Staphylococcus aureus. J. Bacteriol. 187, 5486–5492 (2005). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Delauné A., et al. , The WalKR system controls major staphylococcal virulence genes and is involved in triggering the host inflammatory response. Infect. Immun. 80, 3438–3453 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.A. Fait, Y. Seif et al., Adaptive laboratory evolution and independent component analysis disentangle complex vancomycin adaptation trajectories. European Nucleotide Archive (ENA). https://www.ebi.ac.uk/ena/browser/view/PRJEB52876. Deposited 17 May 2022. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
Sequence data have been deposited in the European Nucleotide Archive under study accession number PRJEB52876 (60) and are publicly available. All other study data are included in the article and supporting information.