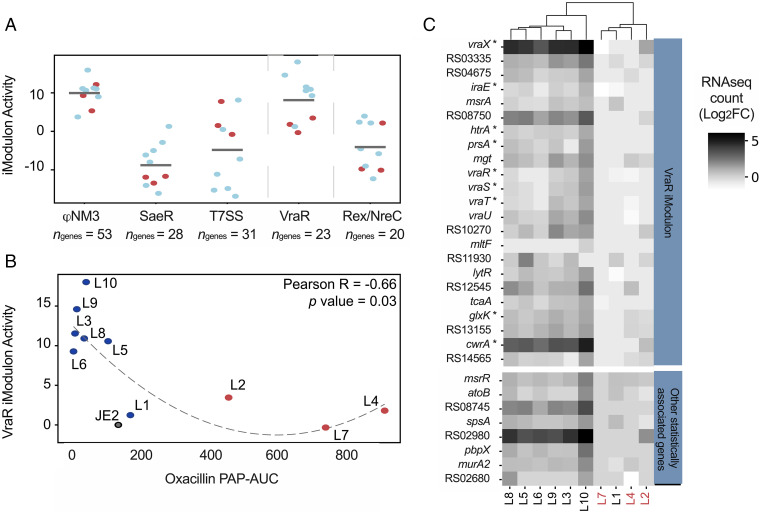
Fig. 5.
Transcriptional profiles related to vancomycin adaptation and β-lactam cross-sensitivity in JE2. (A) ICA of the mutants’ transcriptional profiles reveal five iModulons (independently modulated sets of genes) with the largest changes in activity with respect to wild type. The scatterplot shows the activity levels of each iModulon in each clone. Blue and red dots represent clones with cross-susceptibility and cross-resistance to oxacillin, respectively. (B) The VraR iModulon is the only iModulon showing significant correlation between activity levels in clones and corresponding susceptibility to oxacillin. A decrease in VraR iModulon activity explains increased susceptibility to oxacillin. (C) Cluster map of the expression levels (log2 fold change [FC] with respect to wild type) in the VraR iModulon genes. Genes showing correlated expression levels with MIC are annotated with a star. Additional genes not forming part of the VraR iModulon but that nonetheless also reveal significant correlation with MIC levels are shown separately in the bottom heatmap. Clones are color coded according to cross-resistance to oxacillin (red = lineages with high-oxacillin MIC).